Hibernating ribosomes tether to mitochondria as an adaptive response to cellular stress during glucose depletion
Posted on: 15 November 2023 , updated on: 27 November 2023
Preprint posted on 8 October 2023
Are ribosomes guarding mitochondria during starvation? The fascinating cryo-EM/ET images of ribosomes attached to mitochondria provided in this preprint suggest that they might be!
Selected by Barbora KnotkovaCategories: molecular biology
Background
During nutrient starvation, cells have limited energy to spare and therefore they shut down most anabolic processes. Only proteins whose function will help the cells overcome starvation will be translated under such conditions [1].This leaves the cell with many inactive ribosomes, which are targeted for degradation [2, 3]. However, a recent study examining septin assemblies in the fission yeast Schizosaccharomyces pombe (S. pombe) noted that cytosolic ribosomes accumulate on mitochondria in glucose-starved cells, but the authors did not explore this curious finding further [4]. Gemin and colleagues have now made this phenomenon the main subject of their research and present their findings in this preprint. They illuminate the molecular details underlying the sequestration of inactive ribosomes to mitochondria using cryo-electron microscopy (EM) methods in conjunction with biochemical tools.
Key findings
- Ribosomes enter a self-inhibition mode upon prolonged glucose depletion
-
- After 7 days of glucose depletion of S. pombe, no active ribosomes could be observed by polysome profiling. In addition, no mRNA or tRNA was found to be associated with purified ribosomes – as analysed by single particle cryoEM – suggesting that indeed no protein synthesis was taking place.
- The cryoEM structures of ribosomes from glucose-deprived yeast revealed that the P-site, where the peptide-bound tRNA usually interacts with the ribosome during protein synthesis, is blocked by a helix of the large ribosomal subunit. Furthermore, this conformational shift in the helix disrupts an interaction important for translation initiation.
- Prolonged glucose depletion induces ribosomal tethering to the outer mitochondrial membrane (see Figure below)
-
- Using cryo-electron tomography (cryoET), the research team could analyse the cellular structure of glucose-starved S. pombe. They saw that the mitochondrial network was fragmented, and that the resulting round mitochondria were richly decorated with ribosomes, as has been described before [4].
- To show that ribosomal tethering to the mitochondria is a consequence of glucose-starvation and not mitochondrial fragmentation, the authors inhibited mitochondrial fragmentation in the yeast cells by deleting the mitochondrial fission factor Dnm1. Just like in wild-type cells, ribosomes in Ddnm1 cells associated with the outer mitochondrial membrane upon prolonged glucose depletion, despite the mitochondria being elongated.
- Ribosomes are arranged into oligomers on the outer mitochondrial membrane and face the membrane with the small ribosomal subunit (see Figure below)
-
- The authors analysed the orientation of the ribosomes on the mitochondrial membrane by mapping sub-tomogram averaged electron densities of ribosomes back to the electron tomograms of cell slices.
- The ribosomes were attached to the membrane via the small ribosomal subunit, an orientation distinct from active ribosomes previously found on mitochondria [5, 6].
- The ribosomes formed organised clusters on the mitochondrial membrane with the help of previously unknown binding sites mediating dimer, trimer, tetramer and pentamer formation between ribosomes.
- Ribosomes interact with the outer mitochondrial membrane via the Cpc2/RACK1 subunit
-
- Sub-tomogram averaging of the cryo-tomograms could not provide molecular details about the connection between ribosomes and the outer mitochondrial membrane, probably due to high flexibility. The researchers therefore fitted the single-particle cryoEM structures of purified ribosomes into the tomograms.
- The fit placed the ribosome-associated protein Cpc2 in close proximity to the membrane. Indeed, a Dcpc2 deletion strain did not accumulate ribosomes on mitochondria upon prolonged glucose starvation, suggesting that Cpc2 is the mitochondria-interacting factor of ribosomes.
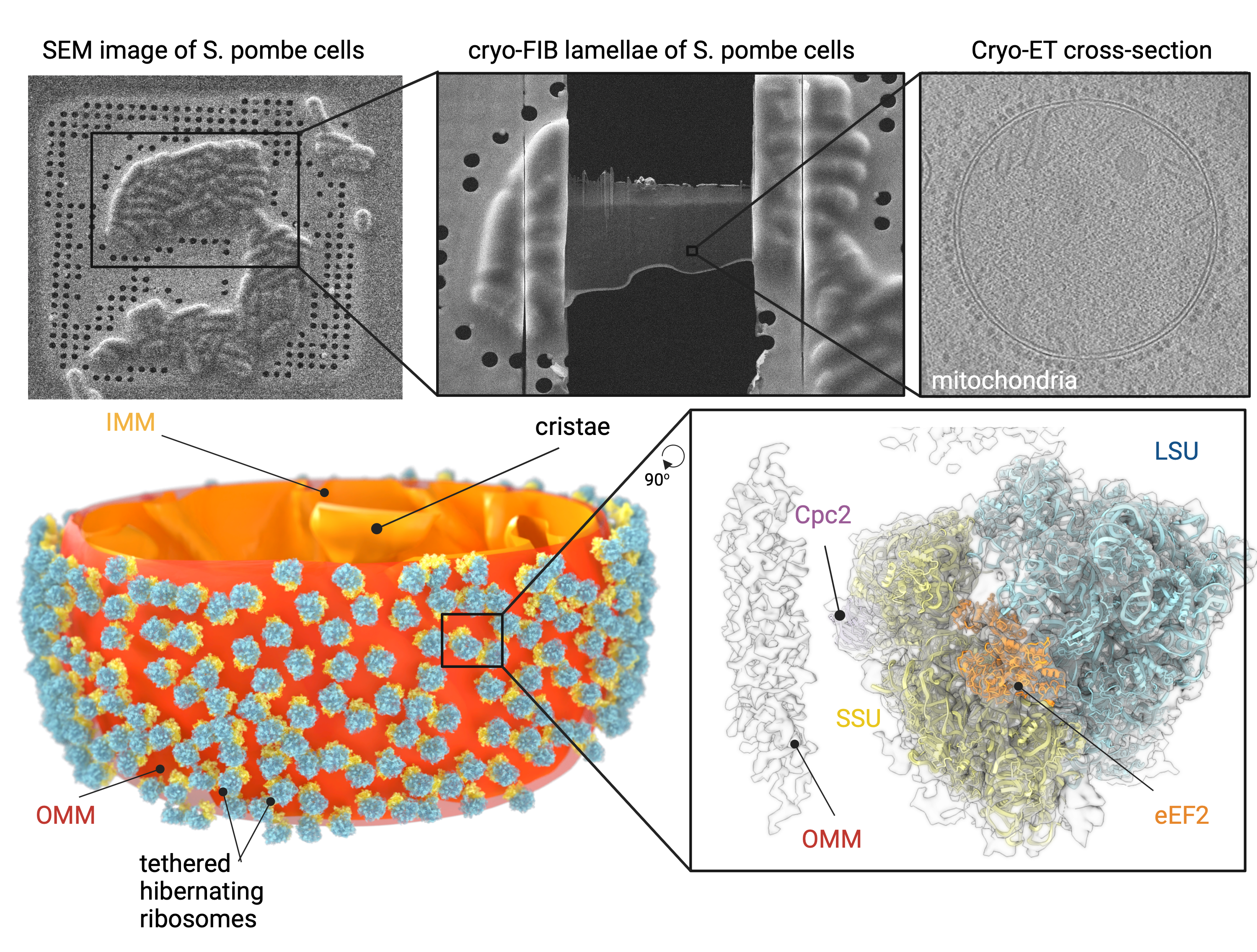
This figure shows how glucose-starved S. pombe cells were sectioned into thin slices by cryo-FIB (A), and how cryoET imaging of these revealed ribosome-decorated mitochondria. Through subtomogram averaging, the mitochondria-tethered ribosomes could be further resolved to determine the location of individual subunits within the ribosome (C). When these higher-resolution ribosomes were mapped back onto a 3D reconstruction of the mitochondrion, all of the ribosomes faced the mitochondrial membrane via their small subunit (coloured in yellow) (D). Figure adapted from the preprint.
Hypotheses for the function of hibernating ribosomes on mitochondria during glucose starvation:
- Ribosomes tethered to fragmented mitochondria protect them from mitophagy and stabilize the mitochondrial potential, thereby averting cell death.
- Mitochondria-bound ribosomes are waiting for the starvation to be over, at which time they can start to synthesise mitochondrial proteins, directly feeding them into mitochondria, which then will have plenty of resources to power the rest of the cell.
What I like about this preprint:
I really enjoyed reading this preprint because it examines such a fascinating phenomenon. I had never heard of ribosomes sitting on mitochondria before and found the cryoET images showing this rather impressive. I also liked that the preprint was not too long and was kept to the point. I hope that there will be more research on this topic and that we can find out more about what these ribosomes are doing on the mitochondria!
Questions and comments
- Do you have any ideas as to what Cpc2’s binding partner in the outer mitochondrial membrane might be? Would it be possible to map translocation complexes of the outer membrane onto the tomograms of mitochondria to determine whether the ribosomes are in close proximity to these, supporting your hypothesis that the hibernating ribosomes are ready to restart mitochondrial protein synthesis upon nutrient repletion?
- Mammalian mitochondria tend to fuse, rather than fragment, during starvation to protect themselves from mitophagy. Do you have an explanation for why pombe mitochondria do the opposite? Do other yeast species’ mitochondria behave the same as S. pombe’s during starvation?
- In your preprint, especially in the abstract, it sounds like you propose that the sequestration of ribosomes to mitochondria may confer cell survival. However, in the corresponding data, Cpc2 deletion impacted cell survival even in EMM 2% glucose media (Figure 4B), where cells are not glucose-deprived, mitochondria are not fragmented and ribosomes are not sequestered to mitochondria (Figure S9). Therefore Cpc2’s interaction with the mitochondrial membrane and the associated ribosome sequestration may not be the factors that confer cell survival, but rather it may be another function of Cpc2 important specifically in the EMM medium but not under glucose-deprivation.
References
1. Janapala, Y., T. Preiss, and N.E. Shirokikh, Control of Translation at the Initiation Phase During Glucose Starvation in Yeast. Int J Mol Sci, 2019. 20(16).
2. Kraft, C., et al., Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol, 2008. 10(5): p. 602-10.
3. An, H. and J.W. Harper, Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol, 2018. 20(2): p. 135-143.
4. Liu, M., et al., Glucose starvation triggers filamentous septin assemblies in an S. pombe septin-2 deletion mutant. Biol Open, 2019. 8(1).
5. Gold, V.A., et al., Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep, 2017. 18(10): p. 1786-1800.
6. Williams, C.C., C.H. Jan, and J.S. Weissman, Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science, 2014. 346(6210): p. 748-51.
doi: https://doi.org/10.1242/prelights.35991
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the molecular biology category:
Cell cycle-dependent mRNA localization in P-bodies
Mohammed JALLOH
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Fetal brain response to maternal inflammation requires microglia
Manuel Lessi
preLists in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (1 votes)
(1 votes)