Real-time visualization of mRNA synthesis during memory formation in live animals
Posted on: 27 October 2021
Preprint posted on 2 September 2021
Article now published in Proceedings of the National Academy of Sciences at http://dx.doi.org/10.1073/pnas.2117076119
Do you remember...? Memory neurons at work: Tracing memory formation in real-time using a genetically encoded RNA indicator
Selected by Kristina Kuhbandner, Soni Mohapatra, Ranabir ChakrabortyCategories: neuroscience
Background
Our brain has an enormous capacity for learning and memorizing, and neuroscientists are only just beginning to understand the mysteries of memory formation and retrieval. The specific neurons involved in memory formation are called memory trace cells or engram cells. These show a characteristic gene expression profile of immediate early genes such as Arc, c-Fos and Egr-1, which are activated transiently and rapidly in response to cellular stimuli. However, conventional methods used to examine immediate early gene transcription, for example RNA fluorescence in situ hybridization (FISH), immunostaining and use of promoter-driven reporter proteins, are limited in terms of temporal resolution and can only provide a snapshot of events (1, 2). In the last few years, researchers have developed a broad palette of optical tools to study neuronal activity during memory formation including genetically encoded indicators for changes in calcium levels or voltage (3).
So far, it has been impossible to continually monitor gene expression in real-time in live animals. In their preprint, Lee et al. address this obstacle by creating a genetically encoded RNA indicator (GERI) mouse line.
Key findings
Development of a GERI to study memory formation
The PP7 bacteriophage coat protein (PCP) binds with high specificity to its cognate PP7 RNA binding site (PBS). The authors employed this PCP-PBS binding system to develop a novel tool for imaging of Arc mRNA. They generated a transgenic PCP-GFP mouse neuronally expressing tandem PCP fused to tandem GFP. Additionally, an Arc-PBS knock-in mouse line where 24x PBS repeats were integrated into the 3’ untranslated region of the Arc gene was created. In the resulting hybrid mouse obtained through crossing the two lines (PCPxArc-PBS), every synthesized Arc mRNA was endogenously labelled with tandem GFP repeats (Fig. 1A). The crossed transgenic mice were subjected to contextual fear conditioning (CFC) wherein animals were exposed to a novel environment and provided with an aversive stimulus (foot shocks). This resulted in fear memory formation, thereby allowing for the investigation of Arc mRNA expression (Fig. 1B). The authors relied on two-photon excitation microscopy of the dorsal CA1 brain region to visualize the labelled mRNA (Fig. 1C). Upon appropriate background correction of the obtained images, a subset of neurons showed bright spots localized in the nucleus, which corresponded to transcription start sites of Arc mRNA (Fig.1D). An investigation of these bright spots through an independent method (two-color FISH using fluorescent probes complementary to the coding sequence and the PBS sequence of the mRNA) confirmed that transcription start sites appear as bright puncta in the acquired images.
In contrast to FISH, this newly developed GERI tool allows for time-lapse imaging of Arc mRNA to study their temporal dynamics. The fraction of neurons with Arc transcription start sites (Arc+) saturated within 7 min after CFC and the signal disappeared 20 min after CFC.
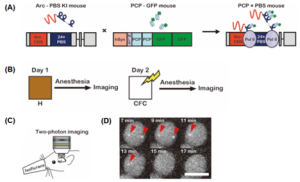
Fig. 1: Imaging of Arc mRNA through a newly developed genetically encoded RNA indicator (GERI) (A) Schematic for in vivo labelling of Arc mRNA with GFP that relies on strong binding specificity between PCP (PP7 bacteriophage capsid protein) and PBS (PP7 binding site) RNA stem-loop. (B) Experimental scheme for imaging of Arc mRNA upon contextual fear conditioning (CFC). (C) Experimental setup for two-photon imaging of Arc mRNA through a hippocampal window (D) Representative time-lapse images of Arc+neuronal cells after CFC. The red arrowheads mark the transcription sites of Arc mRNA (Taken from Fig. 1 of the preprint, made available by CC-By 4.0 license)
Dynamics of Arc-expressing neurons in different brain regions during recent memory retrieval
Next, the authors analyzed changes in Arc-expressing neuronal populations in two different brain regions, the hippocampal CA1 and cortex, during memory encoding and retrieval in the context of fear. Mice were conditioned in a specific context and then repeatedly exposed to this context on three consecutive days to retrieve fear memory. In the CA1 region, 14-20% of the neurons expressed Arc which remained constant during the retrieval trials, whereas in the cortex about 24% of the neurons transcribed Arc with steady increase during the retrieval trials. Interestingly, in the latter area, the group of neurons continuously transcribing Arc during conditioning as well as retrieval was notably higher and the overlap percentage of Arc+ neurons was significantly greater than expected by chance even in a two-day interval trial.
Concordant observations were made over a period of four weeks confirming that repeated reactivation of the same Arc+ neurons was more likely in the cortex than in the CA1, where a nearly random group of cells transcribed Arc following re-exposure to a specific context. In other words, CA1 Arc-expressing neurons seem to have a faster turnover, while in the cortex the Arc+ population is more stable.
Dual imaging of GERI and a calcium indicator
Having established the dynamic nature of Arc+ neurons in the CA1, the authors utilized a dual-color imaging (of Arc and Ca2+) paradigm to analyze the activity pattern in these cells. PCP-PBS mice were injected with an adeno-associated virus expressing jRGECO1a (a red fluorophore for Ca2+ detection) in the dorsal CA1 region. Awake animals were head-fixed and allowed to move on a spherical treadmill whilst navigating a virtual reality (VR) track. These mice were exposed to novel context A for the first two days, and a new context on the third day. Baseline level of Arc was imaged prior to VR exposure, followed by Ca2+ imaging during active navigation, and Arc imaging in anaesthetized animals. VR navigation significantly increased the fraction of Arc+ neurons across the three days of exposure, without impacting the calcium activity. However, on average, neurons that expressed Arc were more active than Arc-negative cells (Arc-), as was evident from calcium activity, burst and theta-burst rates. Activity was consistently higher in cells that expressed Arc on the first two days of exposure (Arc++) compared to Arc– cells. This suggests the involvement of neuronal Arc++ subpopulation in contextual memory.
To determine the overlap between Arc expressing cells and place ensembles upon different context exposures, the authors performed spatial correlation between these groups. Arc+ neurons on day 1 had weak correlation with place cells and significantly lower spatial information than Arc– neurons. This indicates Arc+ neurons act as contributors to episodic memory, rather than storing spatial information. Measuring the Ca2+ activity in different subpopulations of Arc-expressing cells revealed that Arc++ cells had higher computed theta-burst activity in context A. Such activity patterns were also observed in cells expressing Arc on days 2 and 3 in context B. These results are indicative of contextual memory storage in context-specific subpopulations of neurons. Arc++ neurons also had a greater extent of correlation with activity of CA1 neurons during not only memory encoding and storage, but also its retrieval.
Why is this paper interesting?
Memory and its formation has always been a fascinating topic, but unravelling the underlying mechanisms is very challenging. Despite the recent development of new tools, analyzing neuronal activity based on RNA transcription has been limited given that continuously studying RNA expression in real-time has not been feasible. This newly developed GERI by Lee et al. allows easy tracking of RNA expression and transcription start sites over time, without requiring fixing and staining of cells. This RNA imaging technique might facilitate major advances in the field of memory formation and provides virtually endless opportunities to study memory formation in different paradigms. For example, it can be combined with various other tools, such as biosensors or optogenetic tools, or with other mouse models to study pathological changes in memory formation in the context of aging or neurodegeneration, for example in Alzheimer’s disease.
Questions to authors
- Are you also planning to use this technique to look at other mRNAs related to memory formation such as c-Fos or Egr-1? Do you plan on multiplexing GERI with different fluorophores for simultaneous visualization of different mRNAs?
- Would different sensory modalities (odor or noises) have a differential impact on the location and number of Arc+ neurons?
References
(1) Tonegawa, S., Morrissey, M.D. & Kitamura, T. The role of engram cells in the systems consolidation of memory. Nat Rev Neurosci 19, 485–498 (2018). https://doi.org/10.1038/s41583-018-0031-2
(2) Sauvage, Magdalena, Takashi Kitsukawa, and Erika Atucha. “Single-cell memory trace imaging with immediate-early genes.” Journal of neuroscience methods 326 (2019): 108368
(3) Lin MZ, Schnitzer MJ. Genetically encoded indicators of neuronal activity. Nat Neurosci. 2016;19(9):1142-1153. doi:10.1038/nn.4359
doi: https://doi.org/10.1242/prelights.30884
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the neuroscience category:
Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Muhammed Sinan Malik
Post-translational Tuning of Human Cortical Progenitor Neuronal Output
Jawdat Sandakly
Maturation of the glymphatic system confers innate resistance of the brain to Zika virus infection
Jimeng Li
preLists in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)