Structural venomics: evolution of a complex chemical arsenal by massive duplication and neofunctionalization of a single ancestral fold
Posted on: 14 December 2018 , updated on: 17 December 2018
Preprint posted on 3 December 2018
Spider venom disentangled: an interdisciplinary study reveals the complex composition of the Australian funnel-web spider venom and the large contribution of peptide families evolved to have a knotted structure
Selected by Tessa SinnigeCategories: biochemistry
Background
Spider venoms are known to contain a mixture of components including proteins and peptides, which have evolved for millions of years to target ion channels and receptors in order to immobilise prey. Many of the venom peptides are disulfide-rich peptides (DRP’s), and most DRP’s studied so far adopt an inhibitor cystine knot (ICK) fold in which several disulfide bonds create a knotted structure. These so-called ‘knottins’ have the advantage of being very stable and protease resistant. DRP’s can be highly variable in sequence outside of the cysteine residues required for the disulfide bonds, which has made it challenging to map the evolutionary process that gave rise to their striking diversity1.
Results of the preprint
In order to gain insights into the evolutionary history of DRP’s, the authors used a multi-omics and structural biology approach to study the venom from the Australian funnel-web spider Hadronyche infensa. Using MALDI and Orbitrap mass spectrometry on the crude venom they identified 3051 unique peptides, revealing a larger complexity than reported previously for different funnel-web spider species2,3. The authors note, however, that this could be due to improvements in sensitivity of the techniques. They then examined the venom composition in detail at the transcriptome level, and annotated and categorised the sequences to reveal 26 families of DRP’s, in addition to 7 protein families. Proteomics on trypsin-digested venom subsequently confirmed the presence of the majority of the predicted peptides and proteins.
The authors compared the features of the 26 identified DRP families to known structures and sequences, and could assign 12 of them to be knottins with a predicted ICK fold. Another 6 families were definitely not knottins based on homology to other known sequences, leaving 8 families of unknown structure and biological activity. The authors set out to solve the structures of representative DRP’s from these families by NMR spectroscopy, and were able to solve three structures that turned out to be knottins, albeit with different structural elaborations around the typical ICK fold. A fourth structure, on the other hand, represented a completely novel venom peptide fold. Figure 2 of the preprint shows all of the superfamilies with representative structures. The importance of the knottin DRP’s was illustrated by examining the relative expression levels of all the superfamilies now identified as such, adding up ~91% of the total venom peptidome.
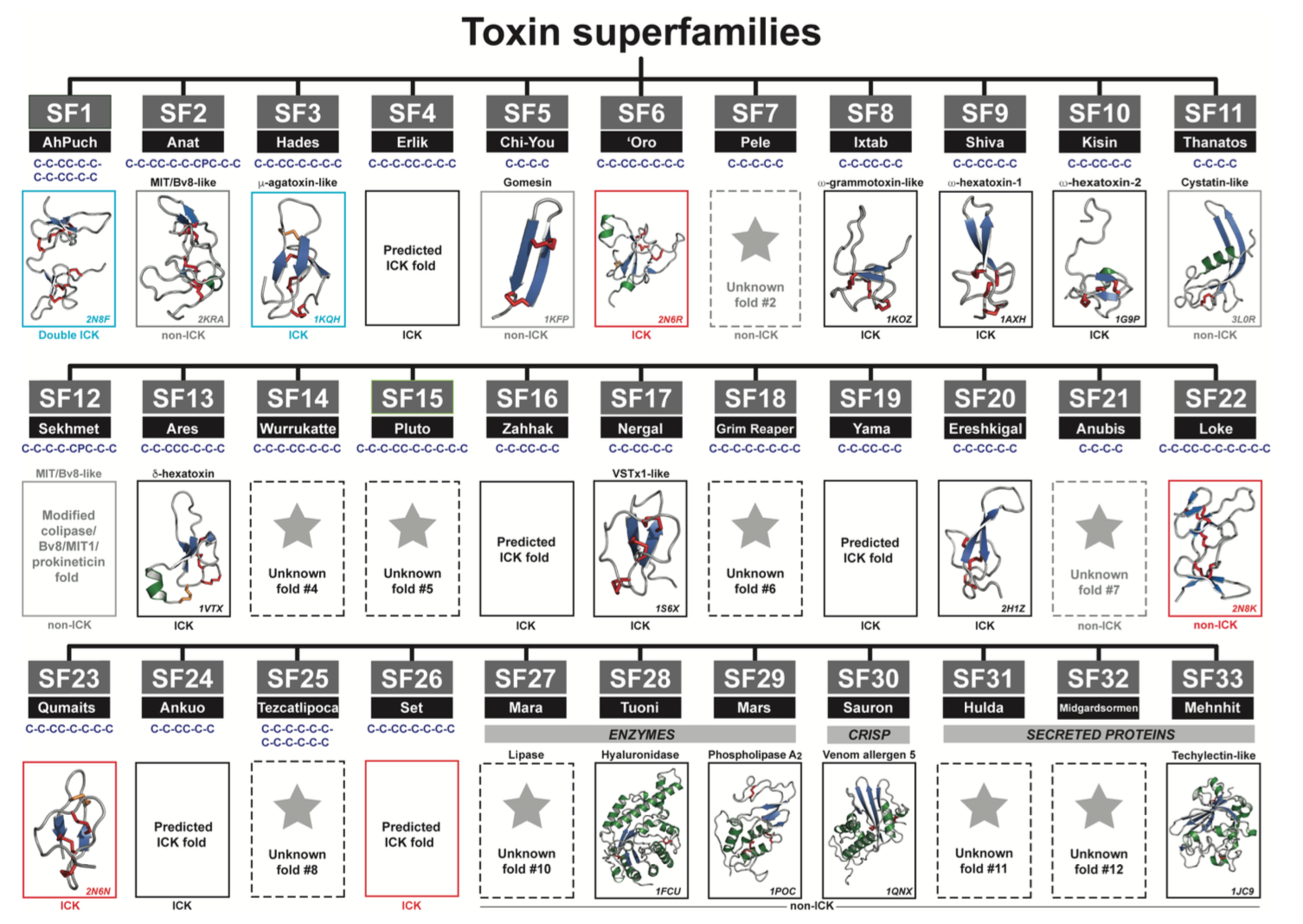
Phylogenetic analysis suggested that the knottin superfamilies likely evolved from the same ancestral fold, although the precise phylogenetic relationship between the different families remained unclear. The phylogenetic tree indicated that either the fourth disulfide bond present in some families evolved independently on at least two occasions, or the ancestral fold contained four disulfide bonds, one of which was later on lost in a subset of families. The analysis showed conclusively that a gain of disulfide bonds occurred for some of the larger peptides, which acquired structural extensions around the knottin scaffold, ruling out that they resulted from gene duplication.
The preprint finally also provided some interesting insights into venom production. When the spider glands were depleted from venom by electrical stimulation, the gene expression pattern changed dramatically when examined three days later, now showing a large fraction of genes involved in cellular processes such as transcription and translation, as well as factors required for protein folding and particularly for the efficient formation of disulfide bonds.
What I like about this preprint
Biology never ceases to amaze me, and I was very excited to learn about the complexity of spider venom and the ways this might have evolved. Furthermore, it is great that the authors went all the way from proteomics and transcriptomics to detailed structure determination with really fascinating results – you don’t discover a bunch of new protein folds every day!
Questions
- First I have a small technical question: what could explain the limited overlap between the peptides identified with MALDI and Orbitrap, respectively?
- What would be required to map the phylogenetic relationships between the DRP superfamilies in more detail? Would it help to have more homologous sequences from other spider species?
- To what extent did the venom peptides co-evolve with their targets?
- With respect to therapeutic implications, could the knowledge about the composition and structural basis of spider venom be useful to improve antivenoms to treat bites? Or could spider venom peptides themselves have pharmaceutical applications?
References
(1) Rodríguez de la Vega, R. C. (2005) A note on the evolution of spider toxins containing the Ick-motif. Toxin Rev. 24, 383–395.
(2) Escoubas, P., Sollod, B., and King, G. F. (2006) Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon 47, 650–663.
(3) Palagi, A., Koh, J. M. S., Leblanc, M., Wilson, D., Dutertre, S., King, G. F., Nicholson, G. M., and Escoubas, P. (2013) Unravelling the complex venom landscapes of lethal Australian funnel-web spiders (Hexathelidae: Atracinae) using LC-MALDI-TOF mass spectrometry. J. Proteomics 80, 292–310.
doi: https://doi.org/10.1242/prelights.6431
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)