Susceptible bacteria survive antibiotic treatment in the mammalian gastrointestinal tract without evolving resistance
Posted on: 16 May 2023
Preprint posted on 11 January 2023
A new model for antibiotic treatment research uncovers an unorthodox method for bacterial survival of treatment. preLight Authors: Hannah Brooks & Derek Resio
Selected by UofA IMB565Categories: genetics, microbiology
preLight Authors: Hannah Brooks & Derek Resio
Background
Antibiotic resistance is the evolution of bacterial mechanisms that protect them from drug treatments. Antibiotic resistance is also a critical component in the overall antimicrobial resistance crisis. With the World Health Organization declaring antimicrobial resistance a top ten global public health threat (1), the importance of elucidating the mechanisms that confer antibiotic resistance is increasingly recognized. Doing so will ideally lead to the development of pharmaceutical treatments that target current drug-resistant microbes and prevent future drug-resistant infections. Since the ingestion of antibiotic-resistant bacteria from animal and food products is a major cause of human infections (2), the mammalian gut is one of the most ideal model systems for studying antibiotic resistance in bacteria. However, one of the issues in antibiotic resistance research is using interval antibiotic administration (intraperitoneal, intravenous, or oral) for research in mice. The use of these methods does not properly mirror antibiotic serum/plasma levels in humans because mice have different antibiotic pharmacokinetics, which can complicate the interpretation of research that uses these methods. Thus, improvements in this area may yield higher quality data.
Typically, bacterial survival during antibiotic treatment is thought to be dependent on resistance, tolerance, or persistence. Resistant bacteria have an inherited ability to grow at high concentrations of an antibiotic regardless of treatment duration, with the concentration at which a bacteria becomes susceptible to treatment being the minimum inhibitory concentration (MIC) (3). Tolerant bacteria, meanwhile, have either an acquired or inherited ability to resist transient exposure to high antibiotic concentrations without the MIC changing (4). Lastly, persistent bacteria are a sub-population that survive high antibiotic concentration exposure while the majority of the population dies off (5). Identifying novel mechanisms that fall under one of these categories is of the utmost importance.
In this preprint, Rodrigues and colleagues propose and demonstrate a new model to study antibiotic treatment in mice (Figure 1). Additionally, they find that different intestinal tissues harbor different concentrations of antibiotics, and ultimately reveal a potential mechanism by which bacteria of the gut can survive antibiotic treatments. Their work highlights the complexity of antibiotic treatments and emphasizes the importance of understanding the mechanisms of antibiotic resistance evolution.
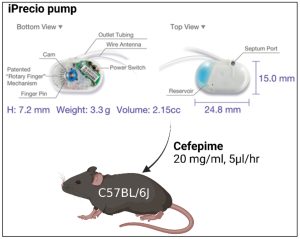
Figure 1: Murine model for continuous antibiotic administration. Implantation of an iPrecio® SMP310R pump allows for continuous administration of cefepime. Cefepime is loaded at 20 mg/mL into the reservoir and administered at flow rate of 5 μl/hour.
Key Findings of this Preprint
Cefepime administration via subcutaneous pump better mimics human pharmacokinetics/dynamics.
Current methods of antibiotic administration in mice that use interval dosing rarely mirror antibiotic plasma levels in humans (6-8). With this in mind, the authors developed a model to continuously administer cefepime subcutaneously. Using these implanted pumps, the researchers could reliably detect cefepime in the plasma, colon, small intestine, and fecal material of the mice. The concentration levels measured in each sample were greater than the minimal inhibitory concentration for the bacteria used in this study, E. coli, which allowed the researchers to properly assess the development of antibiotic resistance. Importantly, the plasma levels of cefepime in mice (~ 5 µg/mL) were within the range of human plasma cefepime treatment levels (~ 2-6 µg/mL) as well. Thus, the researchers developed a suitable model that better mimics human cefepime pharmacokinetics/dynamics and could be used throughout the rest of this study.
E. coli seek refuge in intestinal tissue and have decreased population diversity during cefepime treatment.
To study how gut-colonizing bacteria are affected by cefepime treatment, the authors isolated a pan-susceptible E. coli strain from a pediatric stem cell transplant patient, referred hereafter as the PEc (Parental E. coli) strain. They then barcoded the strain to create the PbEc (Parental barcoded E. coli) strain as a means to test viability and lineage track the strain and assess population diversity. Colonizing germ-free C57Bl/6 mice with PbEc and treating the mice with cefepime for 6 days resulted in the ability to detect PbEc colonization levels from fecal matter. Attempts to culture PbEc from colonic and cecal contents failed; however, viable PbEc was cultured from the ileum. Ap possible explanation for these findings would be that cefepime concentrations in the ileum were not at a bactericidal level. Testing this hypothesis showed that while the cefepime concentration in the ileum (0.8 µg/g) was indeed lower than in the cecal contents (6.8 µg/g) and feces (4.5µg/g), it was still higher than the 0.1 µg/mL MIC of PbEc for cefepime. Additionally, the growth rate and cefepime MIC of PbEc isolated from the ileum (and other intestinal tissues) were similar to the controls.
Daily sequencing of the PbEc strain from feces during cefepime treatment yielded interesting, though perhaps unsurprising, results. Genetic diversity in the PbEc strain over the cefepime treatment course decreased, as evidenced by the daily lower number of unique barcodes compared to control treatment. Also, population diversity decreased in the cefepime-treated mice. These findings suggested that PbEc was able to survive cefepime treatment and gain antibiotic resistance without using a traditional mechanism (i.e., drug inactivation, drug efflux, etc.), and that the surviving populations are subjected to a population bottleneck decreasing genetic diversity over time.
Intestinal tissue E. coli show antibiotic persistence in cefepime-treated mice.
To understand how PbEc ileal isolates were able to survive cefepime exposure, the authors screened bacterial isolates from individual mice for phenotypic differences. Upon doing so, they noticed increased translucency in several of the PbEc isolates from cefepime-treated mice. After performing whole genome sequencing of isolated PbEc ileal strains, they found that two isolates (IM5 and IM6) had mutations in the wbaP gene. Focusing on the IM6 strain, which had a single nucleotide deletion in wbaP, they saw that the IM6 strain had no significant antibiotic resistance compared to PbEc. This led the authors to investigate whether IM6 show an antibiotic tolerant or persistent phenotype. They found that although the minimum duration for killing (which measures how long it takes for 99% or the initially cultured bacteria to die) was similar for IM6 and PbEc, it took significantly longer for IM6 to have a 99.99% reduction in bacterial cells. This finding suggests that the IM6 strain was able to continue growing despite the presence of antibiotics, which led them to conclude that the IM6 isolates had developed an antibiotic persistence phenotype with cefepime treatment.
Increased invasion of colonocyte cells is seen in E. coli mutants lacking wbaP.
To see if disruptions in capsule production in the clinical E. coli strain could have a role in colonocyte invasion, the authors generated a wbaP deletion mutant of the PEc clinical strain. Using intestinal epithelial cell invasion assays, they were able to culture more of the PEc wbaP deletion mutants from colonocytes cells compared to the PEc strain. They also saw that the IM6 isolate showed an increased ability to invade colonocyte cells compared to PEc. By performing a time-kill assay they found, at sub-MIC concentrations, that IM6 still exhibited higher survival compared to PEc. They concluded that IM6 had two fitness advantages over PEc: IM6 has an increased ability to invade colonocyte cells due to the wbaP mutation, and it has cefepime persistence which allows for increased survival in both intracellular and extracellular environments (9).
Conclusion
Taken together, Rodrigues et al. have developed and implemented a clinically relevant model to study how antibiotics can affect bacteria living in the gastrointestinal tract. This model also provides a way to study bacterial population dynamics within a host GI tract by better mimicking the concentration spectrum of antibiotics as well as other things that a bacteria may encounter in the gut. Potential additional stressors – such as co-infections, diet changes, and autoimmune diseases – could be studied using this model.
The authors have shown one mechanism in which commensal gut bacteria can adapt to exposure to antibiotics (by invading intestinal cells where antibiotic concentrations are reduced) and have shown how this can eventually lead to the development of increased antibiotic persistence. This preprint therefore highlights a way in which bacteria can continue to survive after antibiotic treatment. Ultimately, understanding the potential pathways leading to antibiotic resistance can inspire better informed and more effective treatment plans for patients and result in more positive outcomes in the clinic.
What we liked about this preprint & why we chose it
The clinical relevance of this paper is very powerful. The growing concern about antibiotic resistance has led researchers to dive deeper into understanding the mechanisms of how this resistance develops, but also how commensal bacteria can survive antibiotic treatments. (10) We believe that the model and methods described in this preprint will prove to be helpful in future studies of the gut microbiome and antibiotic resistance. We appreciate the author’s emphasis on the importance of designing clinically relevant models to study bacterial resistance and persistence within the gastrointestinal tract. This allows for a more holistic picture of bacterial interactions and could ultimately lead to better treatment plans and better patient outcomes.
References
- https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance#cms
- Verraes C, Van Boxstael S, Van Meervenne E, Van Coillie E, Butaye P, Catry B, de Schaetzen MA, Van Huffel X, Imberechts H, Dierick K, Daube G, Saegerman C, De Block J, Dewulf J, Herman L. Antimicrobial resistance in the food chain: a review. Int J Environ Res Public Health. 2013 Jun 28;10(7):2643-69. doi: 10.3390/ijerph10072643. PMID: 23812024; PMCID: PMC3734448.
- Scholar, E. M. & Pratt, W. B. (2000) The Antimicrobial Drugs. Oxford Univ. Press.
- Brauner, A., Fridman, O., Gefen, O. et al. (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14, 320–330. https://doi.org/10.1038/nrmicro.2016.34
- Gefen, O. & Balaban, N. Q. (2009). The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 33, 704–717.
- Andes, D., and Craig, W.A. (2002). Animal model pharmacokinetics and pharmacodynamics: a critical review. International Journal of Antimicrobial Agents 19, 261–268. doi:10.1016/S0924-8579(02)00022-5.
- Gerber, A.U., Brugger, H.-P., Feller, C., and Stritzko, T. (2022). Antibiotic Therapy of Infections Due to Pseudomonas aeruginosa in Normal and Granulocytopenic Mice: Comparison of Murine and Human Pharmacokinetics.
- Vogelman, B., Gudmundsson, S., Leggett, J., Turnidge, J., Ebert, S., and Craig, W.A. (1988). Correlation of Antimicrobial Pharmacokinetic Parameters with Therapeutic Efficacy in an Animal Model. The Journal of Infectious Diseases 158, 831–847. doi:10.1093/INFDIS/158.4.831.
- Huemer, Markus et al. (2020). Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO reports vol. 21,12 doi:10.15252/embr.202051034
- Baloch, Z., Aslam, B., Khurshid, M., Arshad, M., Muzammil, S., et al. (2021). Antibiotic Resistance: One Health One World Outlook. Frontiers in Cellular and Infection Microbiology. 11. 10.3389/fcimb.2021.771510.
doi: Pending
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genetics category:
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
VANGL2 shapes the mouse heart tube from adjacent epithelia and without planar polarity
Anubhav Prakash
Also in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the genetics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (1 votes)
(1 votes)