Unstructured mRNAs form multivalent RNA-RNA interactions to generate TIS granule networks
Posted on: 8 April 2020 , updated on: 13 April 2020
Preprint posted on 14 February 2020
Unstructured RNAs provide structure to phase separated TIS granule networks.
Selected by Christian BatesCategories: molecular biology
Background
Phase separated bodies, or biomolecular condensates, are a hot topic in current biology after being implicated in an array of diverse and essential cellular functions[1]. These subcellular structures represent regions containing high concentrations of specific biomolecules that exist in a distinct phase: often compared to the droplets that form when oil and water are mixed. Organizing the cell in this way allows transfer of components between the condensate and the surrounding cellular milieu, but not mixing. As such, it is thought that phase separated bodies function to provide order within the crowded cellular environment (Figure 1).
In order to form, these complex phase-separated structures rely on multiple weak interactions, or multivalency. In proteins, this multivalency can be conferred through the presence of promiscuous structured domains capable of binding several partners, or, more commonly, poorly structured low complexity domains. The contributions made by proteins to phase-separation are relatively well understood, and a so-called ‘molecular grammar’ is being developed determining the contribution of specific amino acids to phase-separation capability[2]. Interestingly, in a cellular context these condensates frequently contain large amounts of nucleic acids, such as RNA. The specific RNAs present within phase separated compartments can massively impact the formation of these condensates, modifying key aspects such as viscosity, size and composition[3]. Moreover, testament to its role in phase separation, RNA can even phase separate in isolation. Yet, despite the clear functional importance of RNA in phase separated bodies, the precise properties of RNA that endow it with this ability are less clear and are a topic of great interest.
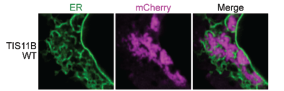
The TIS granule is a recently identified organelle, identified by Christine Mayr’s group in 2018 (Figure 2). This organelle intertwines with the endoplasmic reticulum, a classic and well studied example of a cellular organelle, to promote increased trafficking of specific proteins to the cell surface[4]. However, TIS granules differ from the classical view of organelles as membrane bound subregions of the cell, as it has no membrane and instead is formed by phase-separation. Like many other condensates, TIS granules contain large amounts of RNA, in particular AU-rich RNA. This study set out to understand the contribution made by this AU-rich RNA to TIS granule formation and structure.
Key findings
TIS granules are typified by the presence of the TIS11B protein, an RNA binding protein expressed in a wide range of tissues[4]. As such, the authors began by assessing the impact of mutating the RNA binding domains (RBDs) within TIS11B. Mutating these RBDs altered the TIS granule structure, switching from a mesh- to a blob-like structure. The authors then provide an elegant control, whereby the RBDs from TIS11B were switched for RBDs from a different RNA binding protein that also binds AU-rich RNA, HuR. With this TIS-HuR chimera, the TIS granules reformed into a mesh-like structure again. This suggested that the RNAs bound by these proteins are vital for the appropriate structural formation of the TIS granule.
As both TIS11B and HuR bind AU-rich RNA, the article went on to examine the impact of different RNA species on TIS granule structure. TIS11B alone formed spherical droplets in-vitro, a property that is commonly observed for phase separating proteins. Interestingly, the majority of phase-separated bodies appear as sphere-like structures. In this case then, the TIS granule appears to be the odd one out, by forming a meshwork structure instead. After adding the 3’-untranslated regions (3-’UTRs) from RNAs that are known to localise to TIS granules, TIS11B reverted to the mesh-like structure seen in-vivo. Thus, reaffirming the role of RNA in defining the structure of this condensate. Importantly, the authors controlled for the effects of RNA length and GC content, features that have been described previously to impact condensate formation[5,6].
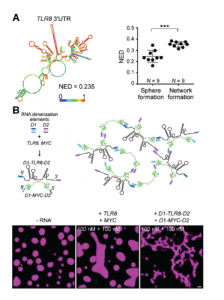
Due to the fact that neither RNA length, or GC content appeared to impact TIS11B network formation, Ma and colleagues next looked at RNA structure using in-silico simulations to predict the probability of base-pairing within the RNA molecule. Ultimately, they arrived at a length-normalized score (NED) for the number of structures predicted for a molecule of RNA. This revealed that RNAs with a more defined structure (a lower NED score) are less likely to induce network formation, and vice-versa (Figure 3A). Perhaps then, unstructured regions of RNA are free to interact with neighbouring RNA molecules through Watson-Crick base-pairing, whereas structured regions have already saturated much of their base-pairing potential by binding to themselves in cis. This seemingly reflects the finding in proteins whereby proteins with disordered regions scaffold and promote phase separation due to their increased multivalency.
Using this information, the paper continued to investigate whether network formation indeed relied on trans-RNA interactions using a deft experiment. Here, the 3’-UTRs from two RNAs that do not normally induce network formation, TLR8 and MYC, were added to purified TIS11B in-vitro. These RNAs are highly structured and are therefore less likely to take part in trans-RNA interactions. Indeed, when added to TIS11B together, these RNAs did not induce network formation. However, when they were artificially ‘stapled’ together through the addition of ‘dimerisation elements’ (sequences of RNA that are the reverse complement of one another), the TIS11B protein displayed the mesh-like structure seen in-vivo. Therefore, under conditions driving trans-RNA interactions, TIS11B adopts to a mesh-like structure (Figure 3B).
Why this work is important
This work builds upon a previous publication from the same lab[4], which defined this novel TIS granule. To date, very little is known about this membraneless organelle, despite the fact that it seems to play a role in and augment important processes such as membrane trafficking. This pre-print looks at TIS granules at the molecular level, determining how the chemistry of biomolecular components drives the unique mesh-like structure that the TIS granule displays. By doing this, it provides not only a more detailed view of this poorly understood cellular compartment, but also a range of tools that can be used in-vivo to further interrogate the function of this granule. For example, it is now possible to use these findings to ask questions about what happens to the cell when the formation of this granule is impeded, or when specific RNAs are excluded from it, by altering RNA structure artificially.
Open questions
- Typically, it is thought that 80S ribosomes, and in particular actively translating ribosomes, are excluded from a range of phase-separated compartments (although a recent pre-print from Jeffrey Chao’s group contends this point[7]). It remains to be proven that translation occurs within this granule (although I may have missed something!); is there any direct evidence for translation at TIS granules?
- TIS granules are thought to be a gel-like condensate, which is typically attributed with slower diffusion rates and a more compact molecular structure. Do you think that if translation does occur within the TIS granule that its biophysical environment is likely to have an impact on the rate of translation?
- Is the mesh-like network that TIS granules display important for their function? Or is it possible that spherical TIS granules would be equally functional?
- Historically phase-separated compartments have been difficult to purify and assess with ‘Omic methods. However, recently a breakthrough in this field has occurred with several labs reporting the purification and interrogation of condensates such as stress granules and P-bodies. Given this, along with the gel-like nature of TIS granules, is it possible to pull down TIS11B and assess the transcriptome and proteome of these TIS granules?
References
- Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
- Wang, J. et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 0, (2018).
- Boeynaems, S. et al. Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. U. S. A. 116, 7889–7898 (2019).
- Ma, W. & Mayr, C. A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3’UTR-Mediated Protein-Protein Interactions. Cell175, 1492–1506.e19 (2018).
- Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017).
- Courel, M. et al. GC content shapes mRNA storage and decay in human cells. Elife 8, (2019).
- Mateju, D., Eichenberger, B., Eglinger, J., Roth, G. & Chao, J. A. Single-molecule imaging reveals translation of mRNAs localized to stress granules. bioRxiv 2020.03.31.018093 (2020).
doi: https://doi.org/10.1242/prelights.18234
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the molecular biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)