Biomimetic, smart and multivalent ligands for G-quadruplex isolation and bioorthogonal imaging
Posted on: 11 January 2021
Preprint posted on 18 December 2020
Article now published in ACS Chemical Biology at http://dx.doi.org/10.1021/acschembio.1c00111
Categories: biophysics
Background:
DNA is a dynamic molecule capable of forming secondary structures beyond its classical double helix appearance. Such structures include G-quadruplexes (G4). When 4 guanine (G) bases in a region of DNA (or RNA) self-associate and bond to each other via Hoogsteen hydrogen bonds (a variation of Watson-Crick base pair bonding which alters the geometry of the DNA tertiary structure), they form a planar G-quartet. When planar G4-quartets stack in the presence of Na+ and K+ ions, they generate G-quadruplex structures which are 4-stranded arrangements of DNA (1). In recent years, key roles for G4 structures have been described in core biological processes such as the regulation of gene expression. Furthermore, their presence has been linked with genomic instability and correlates with cancer predisposition (1,2). Here, the authors generated two small cell-permeable synthetic ligands capable of interacting with and selectively isolating G4 structures from within human cells.
Key Findings:
To generate the ligands synthesised in this study CyTASQ and BioCyTASQ (G4 ligands), the authors designed their compounds based on two previously described reference compounds (PNADOTASQ and BioTASQ; 3,4,5) which are synthetic representations of G-quartet structures (template assembled synthetic G-quartets; TASQs) that have been previously shown to interact with G4 structures in cells according to a biomimetic, like-likes-like approach (that is, the spontaneous association of two G-quartets, one from the native G4 and one from the synthetic TASQ, just like two magnets that attract each other). Here, the authors increased the synthesis yield, reduced the complexity of the chemical reaction required to generate their TASQs and improved compound interactions with G4 structures in cells. A major change the authors made to the chemistry of these compounds was to alter the linker regions between chemical groups within the compounds. They swapped amide links, which provide more rigidity within a molecule, for alkyl links, which increased the flexibility of their compounds.
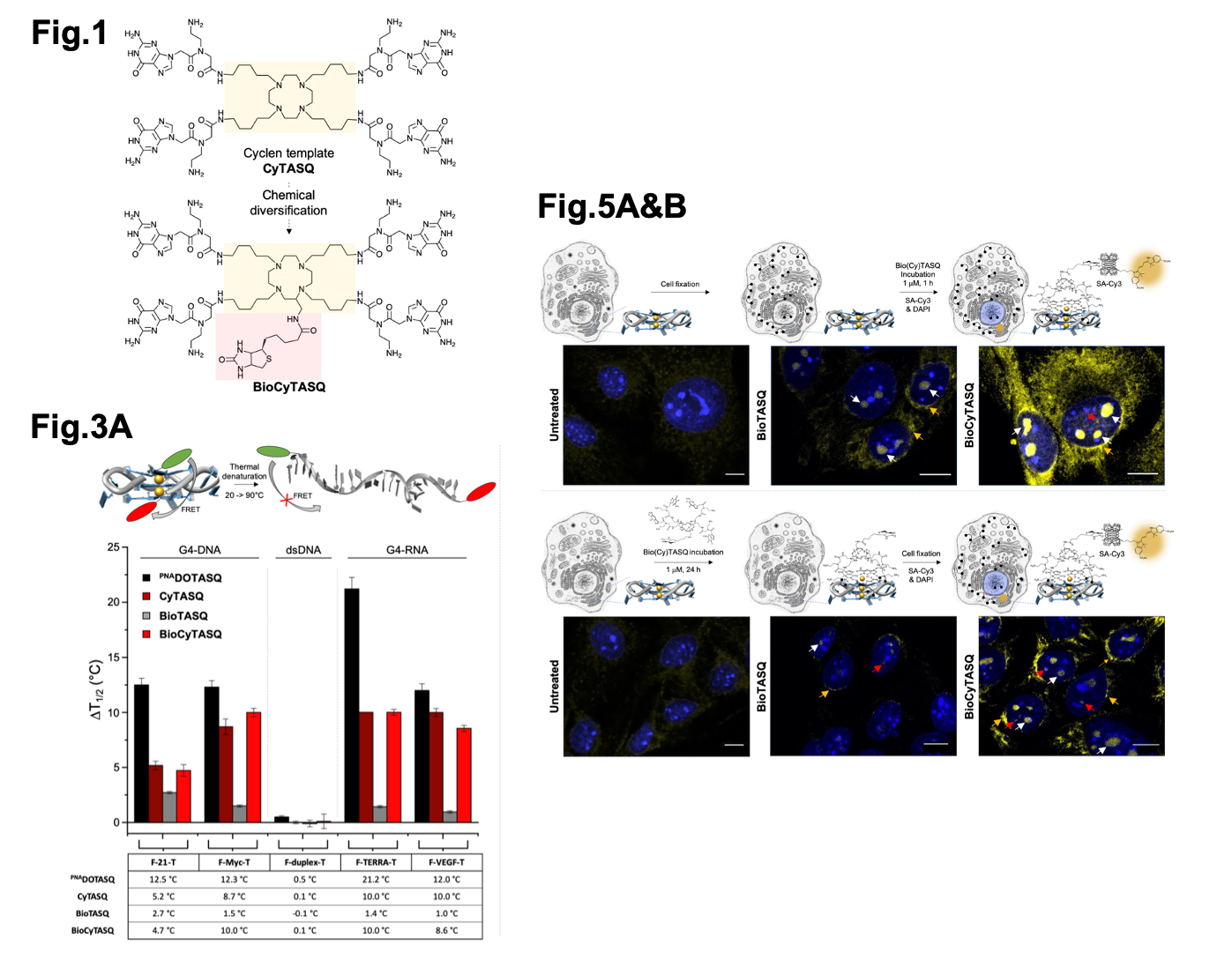
Selected figures from Sperti et al. Fig1 shows the chemical structures of CyTASQ and BioCyTASQ. Fig. 3A shows a schematic of the FRET-melt assay and the corresponding results. Fig 5A & B show the sub-cellular localisation of the TASQ ligands in post-fixed labelled (top) and live cells. Figures modified from Sperti et al. under a CC-BY-NC 4.0 International license.
- CyTASQ and BioCyTASQ interact with G4 structures
To test if CyTASQ and BioCyTASQ interact with DNA/RNA G4 structures, the authors used a Fluorescence Resonance Energy Transfer (FRET)-melting assay (6). FRET-melting is a technique used to examine the physical distance between two fluorescent labels located at both extremities of a G4-forming DNA/RNA sequence. If they are in close proximity, one (the donor) donates energy to the other (the acceptor) resulting in a quenched fluorescence of the donor. If they are far (e.g. upon loss-of-structure DNA melting), this reaction doesn’t happen and the fluorescence of the donor is retained. Here, the authors used doubly labelled synthetic G4s and melted the structures to ask at which temperature half of the signal is restored (which allows for the quantification of the stability of the G4 structures via mid-transition temperatures, T1/2 (in °C)). Then, similar experiments were performed in the presence of their compounds to ask if their compounds influence DNA/RNA structure stability (quantified by a difference of T1/2 values, reported as DT1/2). Overall, their new compounds (CyTASQ and BioCyTASQ) didn’t stabilise the DNA/RNA structures as well as the original compound (PNADOTASQ), likely due to the swapping of the linker regions, but that the biotinylated BioCyTASQ interacted better with G4s when compared to the original BioTASQ (with the amide links) meaning the flexibility in the compound improved its affinity and selectivity for DNA/RNA structures. This was of utmost importance as these biotinylated TASQs were intended to be used as molecular baits to isolate G4s by affinity precipitation (see below).
Of course, in light of their biomimetic nature, the interaction of these compounds was specific to G4 structures.
2) BioCyTASQ can capture G4 structures from synthetic G4 structures in solution
Next, the authors tested the ability of these biotinylated compounds to isolate synthetic G4 DNA/RNA structures (labelled with Fluorescein amidite; FAM) from solution using streptavidin-coated magnetic beads to selectively isolate BioCyTASQ and anything bound to this compound (7). They demonstrate that BioCyTASQ can selectively capture and pull down these synthetic G4 structures (they see increased FAM signal) more efficiently than BioTASQ. These in vitro experiments are important validation steps as biotinylated TASQs are key molecular tools for the G4RP-seq protocol, in which they are used as baits to capture transiently folded RNA G4s in vivo (G4RP: G4-RNA-specific precipitation) prior to their identification by sequencing. The G4RP-seq method thus provides unbiased evidence of the existence of RNA G4s in the transcriptome of functional human cells; the design and synthesis of ever more efficient molecular baits is a key step towards a better understanding of the complex biology of G4s.
3) BioCyTASQ can visualize G4 structures in cells
Another interesting point described here was the versatility of biotinylated G4-ligands. BioCyTASQ was also used to detect G4 structures in human cells by optical imaging. To this end, the authors localise BioCyTASQ after incubation in either live cells or “dead” cells following formaldehyde fixation, by labelling with a fluorescent streptavidin, specifically streptavidin conjugated to Cy3. This protocol, usually referred to as pre-targeted imaging, is applied here to the visualization of G4s in human cells. They saw BioCyTASQ localised in both live and fixed cells diffusely throughout the cytoplasm and at perinucleolar/nucleolus/nuclear regions, where G4s are known to accumulate. This protocol is interesting as it provides an alternative to classical G4 labelling strategies based on G4 molecular probes (8; with potential specificity issues) and the more elegant in situ click imaging (a two-step protocol that circumvents these specificity issues; 9,10).
What I liked about the preprint:
I really enjoyed reading this preprint paper. Despite G4 structures being described in the 1980’s, only recently have we begun to understand their wide roles in eukaryotic cellular regulation. However, we still have much to learn. A current challenge is the selective capture and stabilisation of G4 structures for further study in eukaryotic organisms. In this paper, the authors have used previous data to improve already available compounds generating new compounds suitable for studying and isolating G4 structures selectively in human cells. I really hope to see their use across a range of different organisms in the quest to understand G4 biology.
Questions for the authors:
Q1: The alkyl linkers present in your compounds replace amide linkers (which are more rigid, making the resulting intramolecular G-quartet more stable). By doing this you have increased the ability of BioCyTASQ to selectively interact with synthetic G4 structures over BioTASQ. However, you mention this may alter the conformational stability of the compound. Could you comment on the benefits and drawbacks of using alkyl linkers in place of amide linkers for compound development?
Q2: What are the advantages of using your biotin/avidin G4 labelling approach over in situ click chemistry?
Q3: Does addition of the G4-ligands to live cells alter cell behaviour or cause any cellular toxicity? Have you tested your G4-ligands in any other cell lines i.e are your G4-ligands capable of detecting G4 structures across a range of species and a range of tissue types?
References:
1) Rhodes D., and Lipps, H.J. G-quadruplexes and their regulatory roles in biology. NAR (2015).
2) Varshney, D., Spiegel, J., Zyner, K., Tannahill, D., and Balasubramanian, S. The regulations and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell. Bio (2020).
3) Haudecoeur, R., Stefan, L., Denat, F., and Monchaud, D. A Model of Smart G-Quadruplex Ligand, J. Am. Chem. Soc. (2013).
4) Yang, S. Y., Lejault, P., Chevrier, S., Boidot, R., Robertson, A. G., Wong, J. M., and Monchaud, D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun (2018).
5) Renard, I., Grandmougin, M., Roux, A., Yang, S. Y., Lejault, P., Pirrotta, M., Wong, J. M. Y., and Monchaud, D. Small-molecule affinity capture of DNA/RNA quadruplexes and their identification in vitro and in vivo through the G4RP protocol. Nucleic Acids Res. (2019).
6) Renciuk, D., Zhou, J., Beaurepaire, L., Guedin, A., Bourdoncle, A., Mergny, J-L. A FRET-based screening assay for nucleic acid ligands. Methods (2012).
7) Muller, S., Kumari, S., Rodriguez, R., and Balasubramanian S. Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. (2010).
8) Monchaud, D. Quadruplex detection in human cells. Chapter 5. Ann. Rep. Med. Chem. (2020).
9) Rodriguez, R., Miller, K.M., Forment, J.V., Bradshaw, C.R., Nikan, M., Britton, S., Oelschlaegel, T., Xhemalce, B., Balasubramanian, S., and Jackson, S.P. Small-molecule-induced DNA structures in human genes. Nat. Chem. Bio (2012).
10) Cañeque, T., Muller, S., and Rodriguez, R. Visualizing biologically active small molecules in cells using click chemistry. Nat. Rev. Chem (2018).
11) Moruno-Manchon, J.F., Lejault, P., et al. Small-molecule G-quadruplex stabilizers reveal a novel pathway of autophagy regulation in neurons. eLife (2020).
doi: https://doi.org/10.1242/prelights.26904
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (No Ratings Yet)
(No Ratings Yet)