A deep mutational scanning platform to characterize the fitness landscape of anti-CRISPR proteins
Posted on: 3 September 2021 , updated on: 4 September 2021
Preprint posted on 22 August 2021
Article now published in Nucleic Acids Research at http://dx.doi.org/10.1093/nar/gkae1052
Painting the mutational fitness landscape of proteins through a deep mutational scanning platform
Selected by Soni MohapatraCategories: bioengineering, synthetic biology
Background
CRISPR-Cas systems arm bacteria with a defence mechanism against viruses that prey upon them, also known as phages. Phages, in turn, have evolved counter-defence measures to circumvent CRISR-Cas. Anti-CRISPR (Acr) proteins are small proteins expressed by phages, arising from co-evolutionary dynamics between bacteria and phages (1, 2). Acrs block CRISPR-Cas activity either by inhibiting Cas-DNA binding, by inhibiting DNA cleaving or by preventing proper assembly of crRNA-Cas complex (3, 4, 5). Understanding the structure-function relationship in Acrs, their functional mechanisms and mapping their mutational fitness landscape holds the key to unravelling the secrets of the evolutionary arms-race between phages and their hosts. As well as being of interest to the microbiology research communities, Acrs have been employed to limit off-target editing during CRISPR-Cas based gene editing (6). Engineered Acrs have been integrated to CRISPR-Cas based gene circuits to allow spatial and temporal confinement of gene editing (7, 8). Expanding our knowledge on Acrs can help in designing Acrs with customizable and tunable functionalities for improved gene editing.
Deep mutational scanning (DMS) combines the strengths of next-generation sequencing (NGS) and protein engineering to access the mutational fitness landscape of a protein and develop proteins with novel functionalities and/or better adaptability to different environments (pH, temperature etc.). In this preprint, the authors employ DMS to investigate AcrIIA4 and AcrIIA5, two inhibitors of Streptococcus pyogenes Cas9. They developed a pipeline to map the mutational landscape of these Acr proteins in a high-throughput manner.
Key findings
Establishment of a work-flow for deep mutational scanning of Acr proteins
First, the authors set up a 4-step pipeline for deep mutational scanning of the anti-CRISPR proteins AcrIIA4 and AcrIIA5.
- The first step involves generating a library containing all single point mutants for the chosen Acr proteins through PCR using primer pairs that mutate individual codons with NNB (B = C/G/T) randomized codons.
- The second step requires setting up a phenotype-genotype based screening assay for selection of mutant Acrs that inhibit CRISPR-Cas activity. A sgRNA was designed that facilitates binding of the dSpyCas9-sgRNA to the 5’ region of the RFP reporter non-template strand and blocks expression of RFP. In the presence of a functional Acr mutant that prevents the binding of SpyCas9 to the target DNA, red fluorescence from RFP is observed (Fig. 1).
- For high-throughput screening of functional Acr mutants, the authors adopted a FACS based approach to sort the mutational library based on the intensity of RFP reporter signal reporting for the Cas9 inhibition activity of the mutants. The sorted fractions were then sequenced using NGS.
- A mathematical model was trained using a few benchmark AcrIIA4 and AcrIIA5 mutants and then used to predict the reporter fluorescence distribution for other variants, thereby predicting their Cas inhibition propensities.
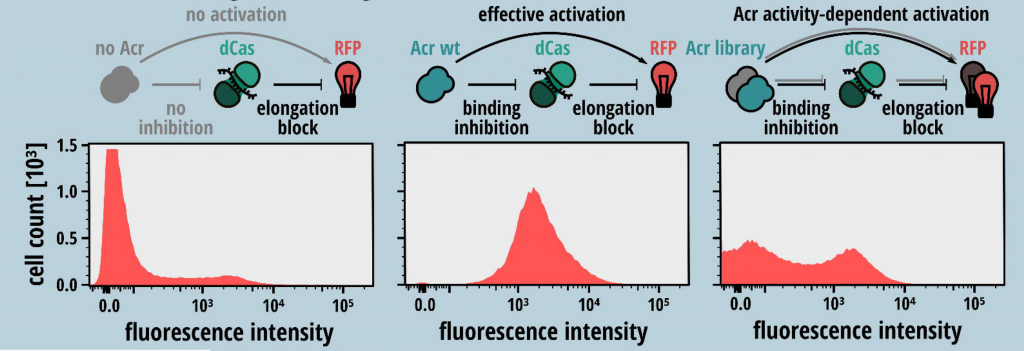
Fig. 1: Schematic of gene-circuit designed for functional screening of Acrs. Higher RFP intensity corresponds to higher inhibitory function of Acr mutants. (Taken from Fig. 1 of the preprint, made available by a CC-BY-NC 4.0 license)
Mapping the mutational fitness landscape of Anti-CRISPR proteins to understand their mutational tolerance
Mapping the mutational landscape of proteins helps us understand which regions of the proteins are tolerant to mutations arising during evolution, identify the functional importance of different protein locations, and understand how different mutations affect the functions of proteins. To construct the mutational fitness landscape of AcrIIA4 and AcrIIA5, the authors adopted the pipeline described earlier. Overall, it was noted that both of these Anti-CRISPR proteins are generally tolerant to occurrences of mutations. The majority (> 70%) of the mutated variants of these Acr proteins retained inhibition potency similar to that of the wild-type proteins. As AcrIIA4 inhibits Cas9 activity through direct interaction with the Cas protein, Acr surface sites that are known to contact Cas were found to be more intolerant to the introduction of mutations compared to other regions of the proteins. For AcrIIA5, mutations in the protein core and in the N-terminal disordered region are less well-tolerated indicating that the intrinsically disordered region of the protein is essential for the inhibitory function of the protein. Interestingly, the mutational library of AcrIIA5 contained variants whose inhibition capabilities are higher than that of the native protein. This pointed towards the possibility of evolutionary development of these better-performing variants, as the native protein is rather weak in its inhibition ability. AcrIIA5 also provides opportunities for better engineered proteins through mutagenesis.
Designing Acr mutants for applications in human cells
The authors demonstrated that their pipeline for selecting Acrs with different inhibition strengths in E. coli is generalizable to other cell types. They adapted their pipeline for human cells by expressing a subset of human codon-optimized AcrIIA4 variants in HEK293T. They relied on a luciferase reporter, a luciferase targeting sgRNA and Cas9 to measure the inhibition activity of Acrs in human cells. A positive correlation was observed between the RFP fluorescence intensities and the bioluminescence signal. At a qualitative level, AcrIIA4 variants when ranked in terms of their inhibition ability were ordered similarly through the E. coli DMS and the human cell assay.
Why I like this preprint
Anti-CRISPR proteins are valuable tools for regulating CRISPR-Cas activity. A deeper understanding of the function of these proteins will allow us to uncover the secrets of gene editing while reducing off-target effects of CRISPR and to engineer anti-CRISPR proteins with higher inhibitory efficiency. In this preprint, the authors set up a pipeline to perform DMS and mapped the mutational fitness landscape of two different Acr proteins, AcrIIA4 and AcrIIA5. Apart from recognizing regions of the proteins that are crucial for the Cas9 inhibitory activity of the proteins, they were able to gain some insights into the functional mechanisms of these proteins. The ability of AcrIIA5 to inhibit DNA binding of Cas9 has been under discussion for a while. The DSM library of AcrIIA5 constituted of some variants whose inhibition capabilities were better than the native protein, suggesting that AcrIIA5 and its mutants are able to inhibit Cas9-DNA binding. Qualitatively, the variants screened through the E. coli DSM platform behaved similarly in human cell lines, signifying the broad applicability of the developed assay.
I believe that this pipeline can be modified for other proteins of interest and used to engineer proteins that allow spatio-temporal control of different cellular processes.
Questions for the authors
- Until now, the ability of AcrIIA5 to block Cas9-DNA binding has been under debate. To resolve this debate, it would be useful to purify AcrIIA5 and its variants that show higher inhibitory function and measure the binding affinity of these proteins to Cas9-RNP. Is that something you are planning to do in the future?
- An earlier article from your lab reported various light controlled variants of AcrIIA4. How do the insertion locations of LOV in AcrIIA4 compare to locations tolerant to mutations obtained through the DSM analysis of AcrIIA4? Could one use the mutational fitness landscape of AcrIIA4 as a guide to determine insertion sites of LOV in the protein?
References
- Bondy-Denomy et al. (2015) Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526,
- Bondy-Denomy et al. (2013) Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429.
- Zhu et al. (2019) Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins. Molecular Cell 74, 296
- Dong et al. (2017) Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein Nature 546, 436
- Harrington et al. (2017) A broad spectrum inhibitor of CRISPR-Cas9. Cell 170, 1224
- Marino et al. (2020) Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nature Methods 17, 471
- Bubeck et al. (2018) Engineered anti-CRISPR proteins for optogenetic control of CRISPR–Cas9. Nature Methods 15, 924
- Aschenbrenner et al. Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity. Science Advances 6, eaay0187
doi: https://doi.org/10.1242/prelights.30466
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)