Double Emulsion Picoreactors for High-Throughput Single-Cell Encapsulation and Phenotyping via FACS
Posted on: 12 August 2020
Preprint posted on 9 June 2020
Article now published in Analytical Chemistry at http://dx.doi.org/10.1021/acs.analchem.0c02499
Dropception: adapting droplet microfluidic analyses to a flow cytometry-compatible format
Selected by Samantha SeahCategories: bioengineering
Background
Traditional droplet microfluidics enables the high-throughput generation of uniformly-sized water-in-oil droplets (1), permitting the production of thousands to millions of independent reaction vessels. This makes it possible to perform over 107 reactions in parallel, while minimising reaction costs. Droplet microfluidics has been used for a plethora of different applications, including single-cell transcriptomic analyses (such as InDrop (2), Drop-seq (3) and 10X genomics (4)), functional antibody screening (5) and the analysis of cytokine secretion (6).
The throughput of water-in-oil droplet generation is extremely high, but the current limiting factor for these droplet microfluidic applications is droplet analysis and sorting. These processes are lower in throughput as compared to flow cytometry and frequently require specialised equipment that are not accessible to non-microfluidic labs. In contrast, flow cytometers are ubiquitous and accessible to most labs.
A recent preprint by Brower and Khariton et al. combine the advantages of droplet microfluidics and flow cytometry. They describe a workflow where single cells are encapsulated into double emulsion (water-in-oil-in-water) droplets that are compatible with flow cytometry, and potentially with fluorescence-activated cell sorting, combining the extensive applications of droplet microfluidics with the high-throughputs and ease of usage of flow cytometry.
Key findings
The authors designed and optimised a workflow for the generation of flow cytometry-compatible double-emulsion droplets containing single cells.
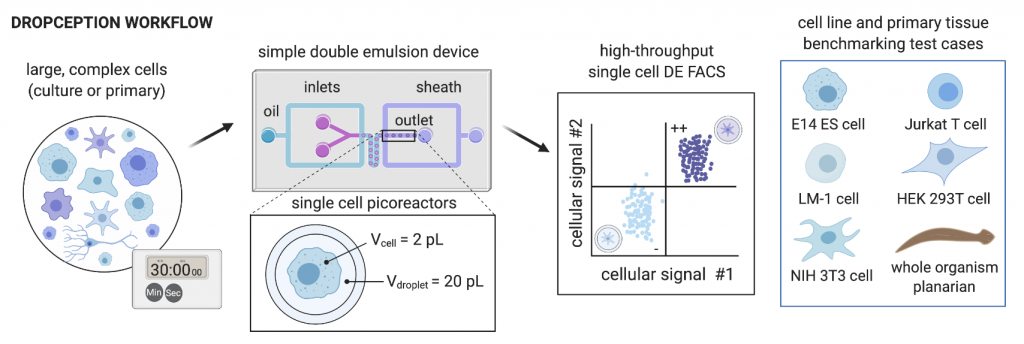
The generation of suitable double emulsion droplets containing cells is not trivial, given that droplets must be small enough to be compatible with analysis by flow cytometry, but large enough to encapsulate mammalian cells, while ensuring that droplets are stable and of a uniform size. This requires identification of optimal droplet-stabilising surfactants, and optimising oil, inner aqueous and outer aqueous flow rates. The authors have done this in their Dropception device, which utilises a dual flow-focusing geometry to encapsulate cells and assay reagents. They produced double emulsion droplets with diameters of 34 and 47 µm for the inner aqueous core and outer oil shell respectively, with high monodispersity (uniformity of size) (2.33% and 1.25% CV on the inner core and outer shell diameters respectively).
The authors then encapsulated single cells into double emulsion droplets. The encapsulation of single cells is governed by Poisson distribution, with many single-cell emulsion techniques employing low cell concentrations (λ < 0.1, where λ is the cell density divided by the droplet volume) to ensure that a majority of cell-containing droplets only contain a single cell. Similarly, the authors carry out single-cell encapsulation with fluorescently-labelled mammalian cells at λ < 0.05 and validated the droplet occupancy with imaging and flow cytometry. The encapsulation and flow cytometry analysis workflow was demonstrated with five different cell types (E14 ES cells, Jurkat, LM-1, HEK 293T and NIH 3T3), with expected occupancies generally observed for all cell types.
To demonstrate compatibility with heterogenous cell populations, such as those present in primary cell populations, the authors encapsulated single cells dissociated from whole planarian flatworms. Even though these single cells vary greatly in size (5-25 μm in diameter), the authors achieved single-cell encapsulation and approached expected droplet occupancies.
Previous work from the same group has demonstrated the reliable recovery of single droplets via single droplet double emulsion FACS (scDE-FACS) based on fluorescence readouts (7). The combination of their previous work, with the single-cell encapsulation workflow outline here, suggests that single cells can be reliably encapsulated into double emulsions, which can then be sorted via FACS. This paves the way for high-throughput single-cell analyses of reaction- or secretion-based phenotypes that are currently impossible with FACS alone.
What I like about this work
I’m a big fan of technologies that are designed and optimised with the user in mind. This technology fits neatly into that category – droplet generation here is relatively simple and requires merely four pumps, while analysis is done with commercially-available and easy-to-use flow cytometers. The workflow is also rapid, and can be completed within 30 minutes to an hour. Overall, I believe that this workflow will revolutionise the field of droplet microfluidics and make microfluidic phenotypic assays accessible to non-specialist labs.
As the authors have mentioned, Dropception is also compatible with various other emulsion-based single-cell technologies, and could enable multi-dimensional analyses. For example, cellular phenotyping made possible with Dropception could be combined with other genomic or transcriptomic analysis, permitting the study of genetic or transcriptomic changes associated with the observed phenotypes. This would permit the expansion of current single-cell assays to include functional, perturbation-based, or multi-omic profiling, greatly increasing the power of future single-cell, and droplet microfluidic analyses.
Questions for the authors
- Do you have any tips and tricks for groups that would like to establish this workflow in their labs? (For example, tricky steps in the workflow that one should take note of.)
- Would further droplet manipulations (merging, splitting etc) be possible with the double emulsion droplets?
- Would it be possible to encapsulate multiple cells within a single double emulsion droplet, and to analyse/sort them with FACS?
Further reading
An earlier work on the same technology by the same group has recently been published in Lab on a Chip. They demonstrate reliable recovery of single droplets via single droplet double emulsion FACS (scDE-FACS) based on fluorescence readouts.
Brower K, Carswell-Crumpton C, Klemm S, Cruz B, Kim G, K. Calhoun SG, et al. Double emulsion flow cytometry with high-throughput single droplet isolation and nucleic acid recovery. Lab on a Chip. 2020;20(12):2062–74. doi.org/10.1039/D0LC00261E
The generation of double emulsions and their sorting by flow cytometry is not new, and has previously been utilised for the screening of bacterial mutant libraries. Please refer to previous publications, for example:
Aharoni A, Amitai G, Bernath K, Magdassi S, Tawfik DS. High-Throughput Screening of Enzyme Libraries: Thiolactonases Evolved by Fluorescence-Activated Sorting of Single Cells in Emulsion Compartments. Chemistry & Biology. 2005 Dec 1;12(12):1281–9. DOI: 10.1016/j.chembiol.2005.09.012
Tu R, Martinez R, Prodanovic R, Klein M, Schwaneberg U. A Flow Cytometry–Based Screening System for Directed Evolution of Proteases. J Biomol Screen. 2011 Mar 1;16(3):285–94. doi.org/10.1177/1087057110396361
References
- Sakai S, Kawabata K, Ono T, Ijima H, Kawakami K. Preparation of mammalian cell-enclosing subsieve-sized capsules (<100 microm) in a coflowing stream. Biotechnol Bioeng. 2004 Apr 20;86(2):168–73.
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015 May 21;161(5):1187–201.
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015 May 21;161(5):1202–14.
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017 Jan 16;8:14049.
- El Debs B, Utharala R, Balyasnikova IV, Griffiths AD, Merten CA. Functional single-cell hybridoma screening using droplet-based microfluidics. Proc Natl Acad Sci U S A. 2012 Jul 17;109(29):11570–5.
- Wen N, Zhao Z, Fan B, Chen D, Men D, Wang J, et al. Development of Droplet Microfluidics Enabling High-Throughput Single-Cell Analysis. Molecules [Internet]. 2016 Jul 5 [cited 2020 Aug 4];21(7). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6272933/
- K. Brower K, Carswell-Crumpton C, Klemm S, Cruz B, Kim G, K. Calhoun SG, et al. Double emulsion flow cytometry with high-throughput single droplet isolation and nucleic acid recovery. Lab Chip. 2020;20(12):2062–74.
doi: https://doi.org/10.1242/prelights.24049
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |











 (No Ratings Yet)
(No Ratings Yet)