Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies
Posted on: 9 May 2019
Preprint posted on 1 March 2019
Article now published in Science Translational Medicine at http://dx.doi.org/10.1126/scitranslmed.aax0876
Programmable bacteria induce durable tumor regression and systemic antitumor immunity
Posted on:
Preprint posted on 1 March 2019
Article now published in Nature Medicine at http://dx.doi.org/10.1038/s41591-019-0498-z
Getting bacteria to do our dirty work: Tal Danino and colleagues develop bacterial platforms to fight cancer.
Selected by Pavithran RavindranCategories: bioengineering, synthetic biology
Background
Major advances in immunotherapy have been made in the past few years. The most recent Nobel Prize in Medicine and Physiology highlighted some of this groundbreaking work wherein James P. Allison and Tasuku Honjo found that cancers cause the down-regulation of our immune system through the use of programmed death ligand 1 (PD-L1) and that targeting this ligand and its receptor, PD-1, can revamp the body’s own immune system to kill the tumor. Analogous research has been done with the CTLA4 receptor. However, these life-saving therapies have many drawbacks: (1) it is still not completely understood why some patients respond and others do not1 and (2) some patients get severe side-effects because of a hyperactive immune system2. For this, researchers have begun to look at so-called “live drugs” where a person’s own immune cells (e.g. T-cells) are engineered to either fight the cancer or to deliver payloads specifically at the cancer, with the hope that these therapies would be much more targeted3.
However, T-cells are highly complex and thus, engineering complex decisions in them is not a simple task. Therefore, these two preprints decided to take an alternative approach: using bacteria. Bacteria provide an ideal platform for such cancer therapeutics as they are relatively simple organisms and synthetic biology has developed a suite of tools to be able to engineer them over the past two decades. Furthermore, research seems to suggest that bacteria are able to selectively localize to cancers due to the immune-protected area that is present in tumor cores4.
The first of these preprints details a synthetic lysis circuit that is used to selectively release nanobodies near tumors and then the second of these preprints utilizes this circuit to release a pro-phagocytic nanobody targeting CD47 that shows remarkable durability.
Key Findings
Gurbatri et al, 2019
In this study the authors set out to create a bacterial based system that would release immuno-therapeutics, specifically a PD-L1 nanobody. As seen in Figure 1 in their design, once the bacteria get to a large enough colony size and reach a quorum threshold, they would signal to each other to lyse and release their internal contents (the PD-L1 nanobody).
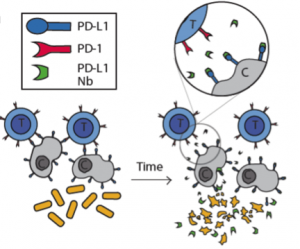
The first thing the authors wanted to confirm was that nanobodies produced in bacteria could indeed block PD-L1. For this, they used flow cytometry on CT26 cells, a cell line that expresses PD-L1. They found that adding just 5% of bacterial lysate containing the PD-L1 nanobody was sufficient to get 50% binding to these cells. From here the authors wanted to carefully characterize the dynamics of the lysis circuit they were going to use in their probiotic where the cells would lyse after reaching a critical density. First they reduced the previously published system to a single plasmid and genomically integrated it such that the bacteria only had one copy. Through simulations, the authors found that with a lower copy number per cell, the bacteria would reach a higher critical density before lysing. In remarkable agreement with their computational model, after using a number of different plasmid vectors to span high to low copy numbers as well their integrated single copy iteration, the authors were able to find exactly this trend in experiments.
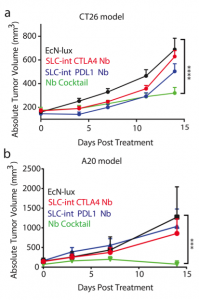
After characterizing the components of their new probiotic, the authors wanted to test their system in a tumor mouse model. They utilized the CT26 cancer line and made a hind flank mouse model from this cell line. The authors then took bacteria expressing PD-L1 nanobody and either their genomically integrated lysis circuit or the circuit driven on a medium copy number plasmid and injected them intratumorally. The authors found that the genomically integrated circuit seemed to work the best as the mice had smaller tumors compared to the mice getting the bacteria with the medium copy number plasmid. The authors took this a step further by introducing bacteria that were producing a nanobody cocktail that inhibit PD-L1 and CTLA-4. They found that this combination therapy was even better at reducing tumor burden in two separate cancer mouse models (figure 2). They found that when the bacteria were releasing the cocktail therapy, the number of regulatory T-cells that suppress the immune response in the tumor was reduced and there was a proliferation of killer T-cells (CD4+). Overall this work was able to generate a bacterial platform that was able to target cancer cells by releasing immunotherapeutics within the vicinity of the tumor.
Chowdhury et al, 2019
In this study, the authors wanted to test the durability of the antitumor response elicited from their bacterial based platforms. In their other preprint, the lab had already established an enhanced synchronized lysis circuit (eSLC) such that once the bacteria grow to a certain density, they lyse and release their contents. In this study, the authors had the bacteria release a nanobody targeting the anti-phagocytic receptor, CD47. Blockade of CD47 should enhance the phagocytosis of cancer cells but also promote the presentation of tumor neoantigens to dendritic cells for sustained immune activation. However, giving systemic blockade of CD47 typically results in anemia as red blood cells also express CD47. The authors first confirmed that the bacteria (a) still undergo synchronized lysis once they hit a critical threshold (b) release CD47 nanobody only upon this lysis and (c) that the nanobody that was produced could indeed block the CD47 receptor.
The authors next wanted to interrogate the clinical efficacy of their nanobody-producing bacteria. They engrafted mice with A20 cancer cells in both hind flanks and each mouse got an injection of one of three treatments: PBS, eSLC expressing bacteria or eSLC expressing bacteria that are producing CD47 nanobody. Mice that received eSLC bacteria did have slightly slower tumor growth compared to PBS, likely due to the activation of the innate immunity response near the tumor. However, the eSLC-CD47 nanobody-expressing bacteria treated mice had near complete regression of their cancer cells. Even more interestingly, when mice were engrafted with a A20 cells, the eSLC-CD47 nanobody-expressing bacteria treated mice survived longer but also were resistant to re-challenge when given new cancer cells again. Mice that only received PBS that got the same re-challenge developed new tumors very quickly. Finally, there were reduced liver and lung metastases in mice receiving the eSLC-CD47 nanobody-expressing bacteria compared to mice getting PBS or eSLC control bacteria. Oddly, no bacteria could be cultured from the liver of the mice getting the bacteria-based treatment, which implies that the bacteria infiltrating the tumor are causing a systemic anti-tumor immunity.
To test this hypothesis more specifically, the authors created a dual-tumor flank mouse model in which cancer cells were engrafted in both flanks of the mice. Only one tumor was treated however with PBS, control eSLC bacteria and the eSLC bacteria producing CD47 nanobody. The authors found that the eSLC-CD47 nanobody-producing bacteria treated mice had reduced growth rates in both the treated and untreated tumors, but also found that the bacteria were not going from the treated tumor to the untreated site. Instead, there may be an adaptive immunity response that is slowing the growth of the secondary tumor.
Overall, the authors developed another probiotic therapy that releases CD47 nanobody once the bacteria have colonized the tumor. Furthermore, they found that this response causes an adaptive immunity response; this is likely due to induction of the systemic adaptation immunity response near the tumor by lysed bacteria as well as abrogation of the CD47 mediated anti-phagocytic activity.
Questions for the authors
(1) Does the PD-L1/CTLA-4 nanobody combination treatment using bacteria also induce the adaptive immunity response as was found in the CD47 treatment case?
(2) As CD47 and PD-L1 seem to be working in very separate mechanisms, would combining PD-L1 with CD47 nanobody bacteria provide a better response compared to the PD-L1/CTLA-4 combination?
(3) Is it better to have two separate bacteria where each produces a nanobody separately or have the same bacterium produce them both?
References
- Rizvi, N. A. et al. Mutational landscape determines sensitivity to PD-1 blockade in non – small cell lung cancer. Science (80-. ). 348, 124–129 (2016).
- Giavridis, T. et al. CAR T cell – induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med. 24, (2018).
- Roybal, K. T. & Lim, W. A. Synthetic Immunology: Hacking Immune Cells to Expand Their Therapeutic Capabilities. Annu. Rev. Immunol. 35, 229–253 (2017).
- Malmgren, R. A. & Flanigan, C. C. Localization of the Vegetative Form of Clostridium tetani in Mouse Tumors Following Intravenous Spore Administration. Cancer Res. 15, 473–478 (1955).
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)