Massively Parallel Selection of NanoCluster Beacons
Posted on: 20 December 2021 , updated on: 23 December 2021
Preprint posted on 5 December 2021
Article now published in Advanced Materials at http://dx.doi.org/10.1002/adma.202204957
In search of a bright beacon: High-throughput screening of nanocluster beacons for new colors and improved brightness
Selected by Soni MohapatraCategories: bioengineering
Background/Introduction
Fluorogenic probes, i.e., probes that fluoresce upon binding to the target, are widely used in different areas of chemistry, biology and medicine. While these probes have diverse uses, the number of such probes are limited owing to difficulties in developing them. Developed in 2010, nanocluster beacons (NCBs) are a class of fluorogenic probes that use DNA-templated silver nano-clusters (AgNC) as reporter and are activated by a proximal G-rich DNA sequence (1). When the activator probe is brought close to the C-rich cluster-nucleation sequence of AgNC, the dark silver clusters become highly emissive. NCBs are easy and cheap to synthesize in a one-pot reaction, show better fluorescent enhancement upon target binding (compared to FRET), and are photo-stable. Recent improvements to NCBs have led to detection of single nucleotide polymorphisms (SNP) in the activator oligo (2,3). However, little is known about how different activator sequences affect the wavelength and enhancement of the fluorescent signal of the reporter. Understanding this would lead to development of better NCBs with improved sensing capabilities.
In this preprint, Kuo et al. set out to investigate the relationship between the activator sequence and the enhancement of reporter signal. They adapted commonly used NGS (next-generation sequencing) chips to screen through a large library of adaptor sequences based on their ability to activate a common probe. Here, the authors describe the development of brighter NCBs of different colors as well as provide a better understanding of how to rationally design better NCBs.
Key findings
High-throughput selection of bright yellow and red NCBs using repurposed NGS chips
The C55 (CCCCCTTAATCCCCC)-templated NC probe lights up when the 18 nucleotide (nt) activator probe, G15 (GGGTGG GGTGGG GTGGGG) is in close proximity to it (Fig. 1(a)). The authors designed oligo libraries of ~40,000 mutated G15 enhancer probes through systematic randomization of nucleobases. With this approach, they sought activator sequences that led to higher enhancement ratio of the common C55 probe compared to the canonical G15 activator sequence. Conventionally, the emission from NCBs upon hybridization with the enhancer probes are measured for one activator sequence at a time. In this preprint, NGS chips (typically used for large scale DNA sequencing) were repurposed to simultaneously image the NCBs activated by a library of ~40,000 enhancer probes in a high-throughput manner. A custom-made bioinformatics and imaging pipeline helps in extracting the sequence of the activators behind each NCB fluorescence spot. This allowed the authors to rank the members of the library from most activating to least activating. Apart from the conventional red NCBs activated by G15, yellow NCBs activated by G12 were also screened. This led to the discovery of activator sequences with higher enhancement ratios for both yellow and red NCBs: yAct4 (TTGGTGGGTGGGGTGGGG, twice brighter than canonical G15) and rAct1(TCCATTGGTGGGGTGGGG, thrice brighter than canonical G12), respectively.
Identification of critical zones in sequences of the activator probes
The 18 nt canonical G15 activator sequence was split into 3 zones of 6 nt each and these three zones were separately randomized (Fig. 1(b)) to investigate the impact of these mutations on NCB brightness. The study revealed that the second zone from nucleotides 7-12 are crucial for the activation of NCBs. Positions 10-12 were found to be especially sensitive to mutations, indicating that nucleotides 7-12 of the activator might be engaged in hybridization with the templated DNA of the AgNC probes for activation.
While the high-throughput screening of different regions of the activator sequence allows for identification of hotspots, it is not trivial to come up with a golden rule to rationally design better activator sequences. The different zones of the enhancer also work cooperatively to enhance the probe signal making it difficult to predict the best activator sequence for brighter NCBs. The authors therefore employed machine learning algorithms trained on the generated big data sets to rationally develop better and brighter NCBs. Through their pipeline, two new activators (yPred1 and rPred3) were discovered. yPred1 had 12 guanines and rPred3 had 9 guanines, lower than the traditional G15 and G12, respectively.
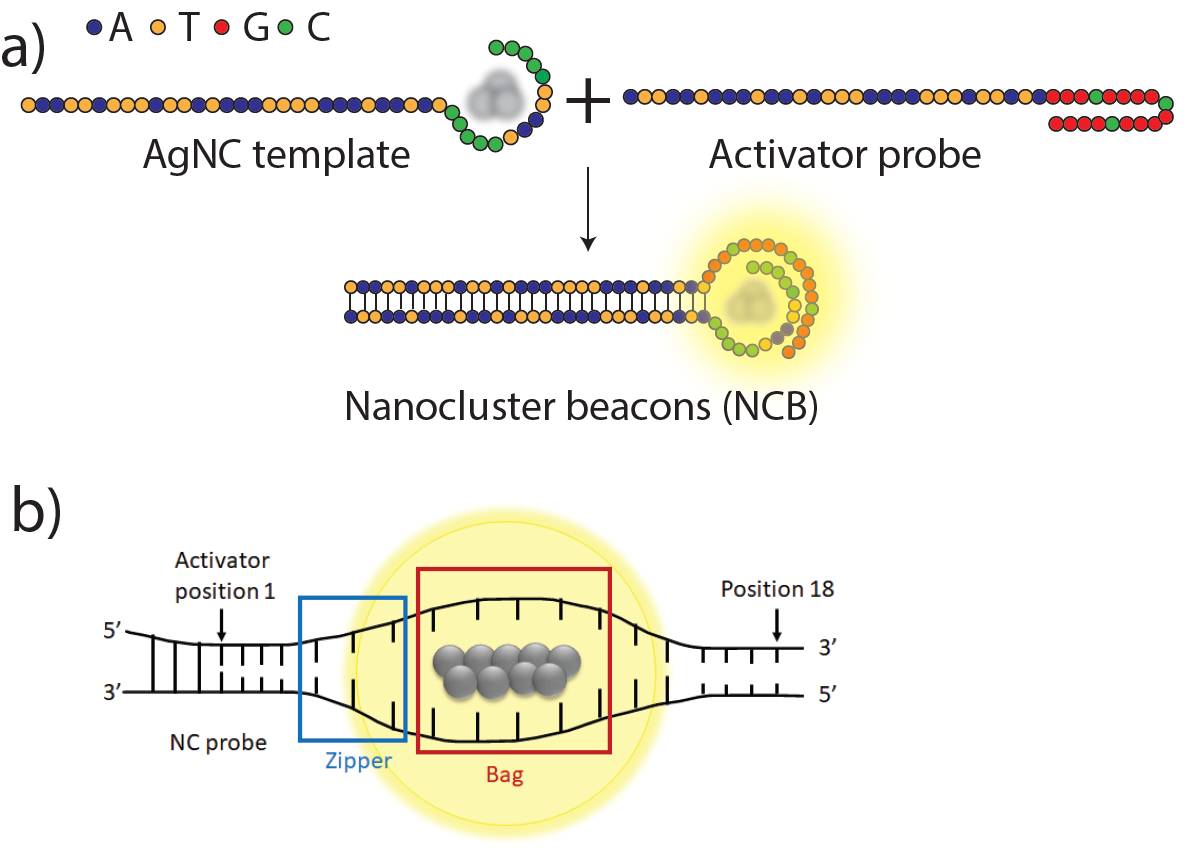
Fig. 1 (a) A DNA-templated silver nanocluster (AgNC) interacts with a guanine-rich proximal activator sequence creating an activated NCB. (b) Schematic of a zipper bag model depicting the nucleotide positions that constitute the “zipper” and the “bag” (Figures adapted from the preprint)
Zipper bag model as a mechanistic explanation for NCBs
Detection of genetic mutations, especially SNPs, that lead to diseases is a challenging problem. In the past, NCBs have been employed for detection of SNPs (2, 3). Usage of NGS chips for massively parallel screening of NCBs offers the benefit of detecting enhancer sequences that lead to largely different enhancement ratios, despite differing by a single nucleotide. In the context of this preprint, POTs (Polar Opposite Twins) are a pair of activators that differ by a single nucleobase but show a drastic difference in brightness of the activated NCBs. Identification of POTs are critical to understanding the mechanistic details of activation of NCBs as well as designing activator sequences for detection of SNPs.
The two extreme POTs showing the most different enhancement ratios that were identified in this work: yPOT5-yPOT6 (yellow NCB) and rPOT5-POT6 (red NCB) were substituted at position 5 (G to C) and position 4 (C to T), respectively. These substitutions were in the first zone of the canonical activator sequence i.e nucleobases 2-6, outside the critical zone of 7-12 nucleotides (Fig. 1(b)). This led to the proposal of a zipper bag model as an all-encompassing explanation of the sequences that activate AgNCs. In the zipper bag model, the 7-12 nucleobase region of the 18 nt activator sequence is the “bag” that holds the NCB chromophore. The zipper region, discovered through identification of POTs, is responsible for sealing the “bag”. Minor changes in the zipper alter the closing of the “bag” and the environment of the chromophore, leading to changes in emission spectrum.
Why I like this preprint
Over the last decade, NCBs have been applied to detect proteins, cancer cells, metabolites to name a few (4-9). Discovery of new and brighter NCBs can further facilitate their application in a wide range of avenues. While previously published work has looked into the impact of activator sequences on the brightness of NCBs, the number of different sequences that can be investigated using conventional test tube-based experiments is very limited. In this preprint, the authors repurposed NGS chips for simultaneous high-throughput screening of ~40,000 activator sequences. Through this, they discovered previously unknown activator sequences that resulted in brighter NCBs, compared to canonical activators. This also led to the identification of the critical regions that are sensitive to changes in nucleotide sequences. The discovery of POTs drove the establishment of the zipper bag model that provides a mechanistic explanation for the activation of NCBs. Their high-throughput screening approach provided the authors with access to a large data set. They therefore utilized machine learning based approaches to predict activator sequences that result in enhancement of emission from NCBs. Some of the promising candidates were tested experimentally and were found to be in qualitative agreement with their prediction. In the future, studies combining high-throughput screening and machine learning algorithms, such as the one reported here, might hold the key to rationally designing NCBs for different kinds of applications.
Questions for authors
- Given the large training data set you have available, is it possible to come up with novel template-activator pairs? It would be interesting to have new/alternative non-C-rich templates that are not in circulation currently.
- Would a truncated G15 or G12 enhancer be enough to hybridize and activate the AgNC?
References
- Yeh, H.C. et al. A DNA-silver nanocluster probe that fluoresces upon hybridization. Nano Letters 10, 3106-3110, doi:10.1021/nl101773c (2010).
- Obliosca, J. M. et al. A complementary palette of NanoCluster Beacons. ACS Nano 8, 10150- 10160, doi:10.1021/nn505338e (2014).
- Yeh, H. C. et al. A fluorescence light-up Ag nanocluster probe that discriminates singlenucleotide variants by emission color. Journal of the American Chemical Society 134, 11550- 11558, doi:10.1021/ja3024737 (2012)
- Yin, J. et al. Label-free and turn-on aptamer strategy for cancer cells detection based on a DNA– silver nanocluster fluorescence upon recognition-induced hybridization. Analytical Chemistry 85, 12011-12019, doi:10.1021/ac402989u (2013)
- Li, J. J., Zhong, X. Q., Zhang, H. Q., Le, X. C. & Zhu, J. J. Binding-induced fluorescence turn-on assay using aptamer-functionalized silver nanocluster DNA probes. Analytical Chemistry 84, 5170-5174, doi:10.1021/Ac3006268 (2012)
- Juul, S. et al. NanoCluster Beacons as reporter probes in rolling circle enhanced enzyme activity detection. Nanoscale 7, 8332-8337, doi:10.1039/c5nr01705j (2015)
- Chen, Y. A. et al. NanoCluster Beacons Enable Detection of a Single N(6)-Methyladenine. Journal of the American Chemical Society 137, 10476-10479, doi:10.1021/jacs.5b06038 (2015).
- Zhang, J. et al. Hairpin DNA-templated silver nanoclusters as novel beacons in strand displacement amplification for microRNAs detection. Analytical Chemistry 88, 1294-1302, doi:10.1021/acs.analchem.5b03729 (2015).
- Zhang, M., Guo, S. M., Li, Y. R., Zuo, P. & Ye, B. C. A label-free fluorescent molecular beacon based on DNA-templated silver nanoclusters for detection of adenosine and adenosine deaminase. Chem Commun 48, 5488-5490, doi:10.1039/c2cc31626a (2012).
doi: https://doi.org/10.1242/prelights.31182
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |











 (No Ratings Yet)
(No Ratings Yet)