Restructuring of an asymmetric neural circuit during associative learning
Posted on: 30 January 2023
Preprint posted on 13 January 2023
The only constant is change – even in “hard-wired” C. elegans. Rewiring of an asymmetric neural circuit leads to associative salt-learning.
Selected by Chee Kiang EweCategories: animal behavior and cognition, genetics
Background:
In 1977, John Sulston and Bob Horvitz traced the cell lineage of C. elegans larvae and found that C. elegans development is highly robust; hermaphrodites contain 1090 somatic cells, 131 of which undergo programmed cell death, leaving invariant 959 cells (Sulston et al., 1983; Sulston & Horvitz, 1977). Around the same time, John White reconstructed the connectome, which consists of 302 stereotyped neurons, using electron microscopy (White et al., 1986). While these classical studies imply that C. elegans development, just as its nervous system, is hardwired, a recent preprint demonstrates that synaptic connectivity may undergo significant restructuring during associative learning. The authors of this preprint could show that salt conditioning alters the architecture of the salt sensing and learning circuit, which is controlled by the conserved insulin signaling (IIS) pathway. This study therefore provides an important insight into the molecular basis for the specification of asymmetric brain functionality.
Key findings:
- Salt exposure leads to rewiring of synaptic connectivity and associative learning in elegans.
As shown previously (Kunitomo et al., 2013), the authors reported that (naïve) worms grown in low salt (33 mM) conditions prefer a low salt environment over a high salt environment in a chemotaxis assay. In contrast, worms grown in high salt (100 mM) conditions for 12 hours tended to chemotax towards agar with a high salt content. C. elegans detects salt using a pair of ASE sensory neurons (left: ASEL; right: ASER). Using a reporter for the presynaptic active zone and iBLINC technology, the authors noted that naïve worms showed a left bias, while conditioned worms showed a right bias in the ASE connection with their postsynaptic partners AWCL/R. The switch in left/right bias in ASE>AWC connection appears to drive differential salt preference, demonstrating that synaptic “hardwiring” is indeed plastic and may change with experience.
- ASE cell fate in part determines asymmetric connectivity.
Transcriptional specification drives left/right asymmetry in both ASE and AWC neurons (Hobert, 2014). Intriguingly, the authors found that symmetrizing ASE, but not AWC, altered ASE>AWC connectivity. Left biased ASE>AWC connectivity was observed when both ASE adopted an ASER fate, as in naïve wildtype; however, a right bias was observed when both ASE adopted an ASEL fate, indicating that while ASE fate may influence left/right asymmetric connectivity, there are other unknown factors at play here since asymmetry persists even when ASE neurons are symmetrized.
- The IIS pathway regulates synaptic connectivity.
It was previously found that the IIS pathway is important for salt sensing and learning (Tomioka et al., 2006). Indeed, the authors showed that knocking out the gene encoding the insulin receptor daf-2 or the downstream PI3 kinase age-1, abrogated asymmetry in ASE>AWC connectivity. Using the Cre-Lox system to achieve cell-specific knockouts, the authors showed that DAF-2 plays a role in establishing asymmetric connectivity in ASE, but not in AWC. Interestingly, removing daf-2 in ASEL in naïve worms led to a reduced connectivity on the left side and a more symmetric pattern. Nevertheless, salt conditioned mutant worms still showed a right-side bias. In contrast, eliminating daf-2 in ASER did not affect connectivity in naïve worms, but conditioned animals showed symmetrized ASE>AWC connections. These results indicate that the IIS pathway promotes synaptic formation and that DAF-2 is asymmetrically activated. Indeed, by examining the localization of DAF-16/FOXO (IIS effector), the authors could show that the IIS pathway is more active in ASEL in naïve worms and in ASER in conditioned worms, thereby restructuring the synaptic connectivity during associative learning.
Next, the authors examined the involvement of insulin-like peptide in ASE>AWC connectivity and found, surprisingly, that worms lacking ins-6, unlike daf-2(-) mutants, showed a right bias and preferred high salt agar. This indicates that while INS-6 is important in regulating ASE>AWC connectivity, the outcome of DAF-2 activation is likely modulated by multiple insulin-like peptides. Interestingly, the authors found that INS-6 was expressed in two pairs of ASI and ASJ sensory neurons. While the expression of ins-6 is symmetric in ASI with or without salt conditioning, it is stronger in ASJL than in ASJR in naïve animals; the reverse is true in conditioned animals, mirroring ASE>AWC connectivity. Intriguingly, eliminating ins-6 in ASJ alone symmetrized ASE>AWC connectivity, whereas removing ins-6 in both ASJ and ASI led to a right bias, recapitulating the ins-6(-) mutant’s phenotype. Finally, the authors knocked out ins-6 specifically in ASJL, but not in ASJR, and observed a right bias in ASE>AWC connectivity. These results indicate that both the expression and the source (ASJ vs. ASI) of INS-6 are important in modulating ASE>AWC asymmetry (Figure 1).
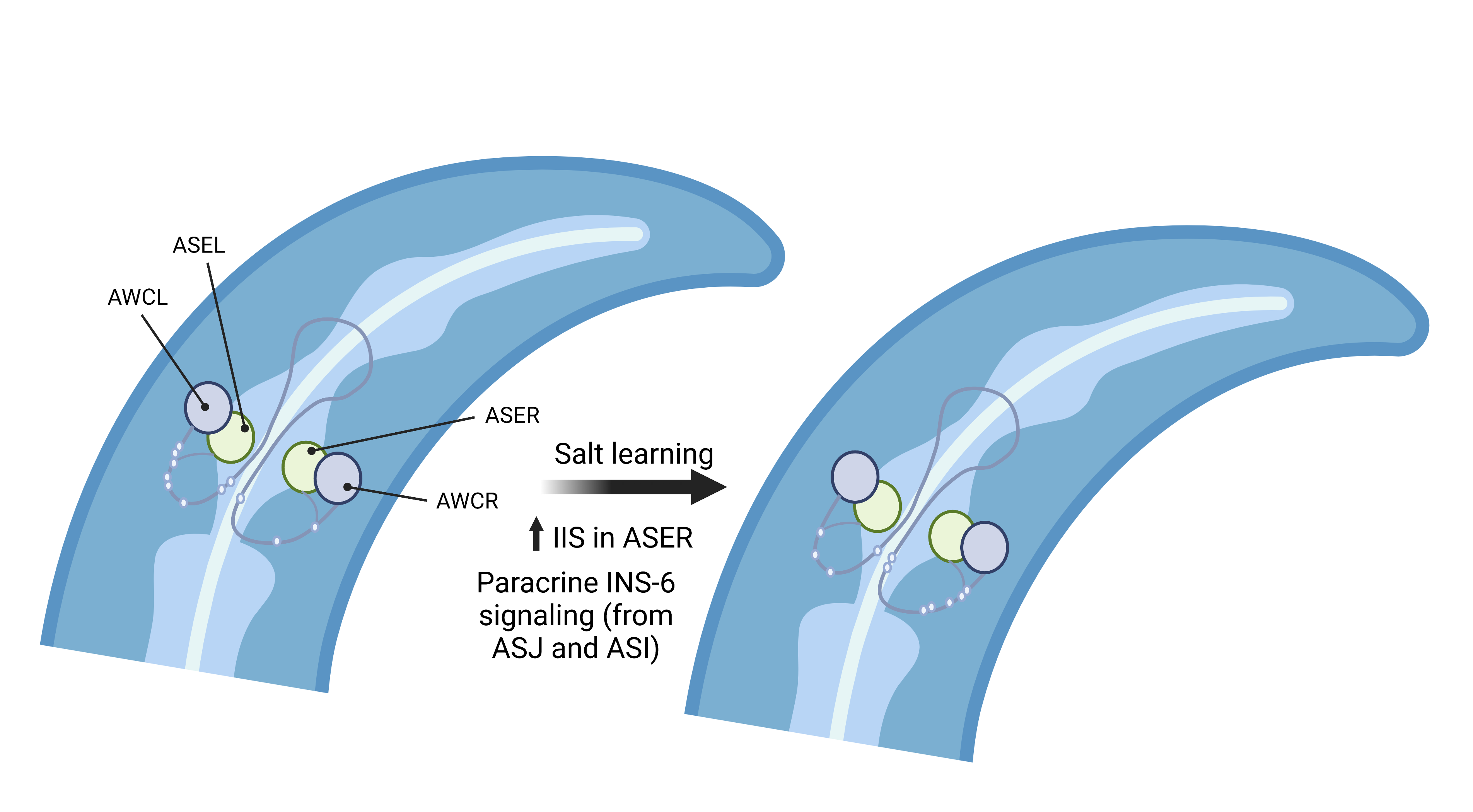
Figure 1: INS-6 and the IIS control the left/right asymmetry of ASE>AWC connectivity (created with BioRender.com).
What I liked about this preprint:
Tang and colleagues beautifully show how asymmetrical activation of IIS leads to associative learning in C. elegans. The experiments are thorough, and I am especially impressed by the clever use of the Cre-Lox system to generate cell-specific knockouts. As the authors have pointed out, the expression of insulin-like peptide in the nervous system is evolutionary conserved, raising the exciting possibility that IIS may also mediate changes in neuronal network architecture in other organisms.
Questions for the authors:
- It is very interesting that knocking out ins-6 in ASJ or ASI gives rise to different phenotypes. How do you think ASE distinguishes INS-6 released from ASJ or ASI?
- Have you tried conditioning the worms at L1 or L2 to see if that affects the establishment of the asymmetric connectivity later in their lives?
References:
Hobert, O. (2014). Development of left/right asymmetry in the Caenorhabditis elegans nervous system: From zygote to postmitotic neuron. Genesis, 52(6), 528–543. https://doi.org/10.1002/DVG.22747
Kunitomo, H., Sato, H., Iwata, R., Satoh, Y., Ohno, H., Yamada, K., & Iino, Y. (2013). Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt concentration chemotaxis in Caenorhabditis elegans. Nature Communications, 4. https://doi.org/10.1038/NCOMMS3210
Sulston, J. E., & Horvitz, H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Developmental Biology, 56(1), 110–156. http://www.ncbi.nlm.nih.gov/pubmed/838129
Sulston, J. E., Schierenberg, E., White, J. G., & Thomson, J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology, 100(1), 64–119. http://www.ncbi.nlm.nih.gov/pubmed/6684600
Tomioka, M., Adachi, T., Suzuki, H., Kunitomo, H., Schafer, W. R., & Iino, Y. (2006). The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron, 51(5), 613–625. https://doi.org/10.1016/J.NEURON.2006.07.024
White, J. G., Southgate, E., Thomson, J. N., & Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 314(1165), 1–340. https://doi.org/10.1098/RSTB.1986.0056
doi: https://doi.org/10.1242/prelights.33580
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the animal behavior and cognition category:
Cannibalism as a mechanism to offset reproductive costs in three-spined sticklebacks
Tina Nguyen
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
Trade-offs between surviving and thriving: A careful balance of physiological limitations and reproductive effort under thermal stress
Tshepiso Majelantle
Also in the genetics category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
preLists in the animal behavior and cognition category:
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Bats
A list of preprints dealing with the ecology, evolution and behavior of bats
| List by | Baheerathan Murugavel |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Also in the genetics category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (1 votes)
(1 votes)