Systematic Engineering of Artificial Metalloenzymes for New-to-Nature Reactions
Posted on: 17 October 2020 , updated on: 4 April 2021
Preprint posted on 11 August 2020
Article now published in Science Advances at http://dx.doi.org/10.1126/sciadv.abe4208
A call to ArMs: researchers develop and screen artificial metalloenzymes for new reactions using a systematic and high-throughput strategy
Selected by Zhang-He GohCategories: bioengineering, synthetic biology
Background of preprint
Artificial metalloenzymes (ArMs) generally exhibit exceptional catalytic performance and selectivity under relatively mild reaction conditions. Boosted by advancements in complementary fields, such as bioengineering and synthetic biology, ArM development has propelled the fields of sustainable synthesis. One commonly used technique to generate new ArMs centres on the use of biotin-streptavidin technology. This strategy relies on the high affinity of streptavidin (Sav) for the vitamin D-biotin to non-covalently anchor the cofactor—biotinylated metal complexes—within the Sav protein.
Currently, the development of ArMs largely relies on whole-cell screening, but this has its own challenges, such as insufficient cofactor uptake into the cell and cofactor poisoning by cellular components. While some of these challenges have been circumvented in specific cases, there have not been any general solutions to this problem to date. To mitigate this challenge, Vornholt et al. have developed a well-plate-based screening protocol. Specifically, the authors demonstrated the general applicability of the protocol to new reaction conditions, cofactors, and analytical methods, and showed that their protocol has the added benefit of being amenable to lab automation as well.
Key findings of preprint
The work by Vornholt et al in this preprint can be divided into three main parts (Fig. 1). First, they constructed and characterised a sequence-defined Sav library. Second, they screened ArMs for five bioorthogonal reactions. Third, they analysed sequence-activity landscapes to understand the correlation between biophysical properties and activity changes.
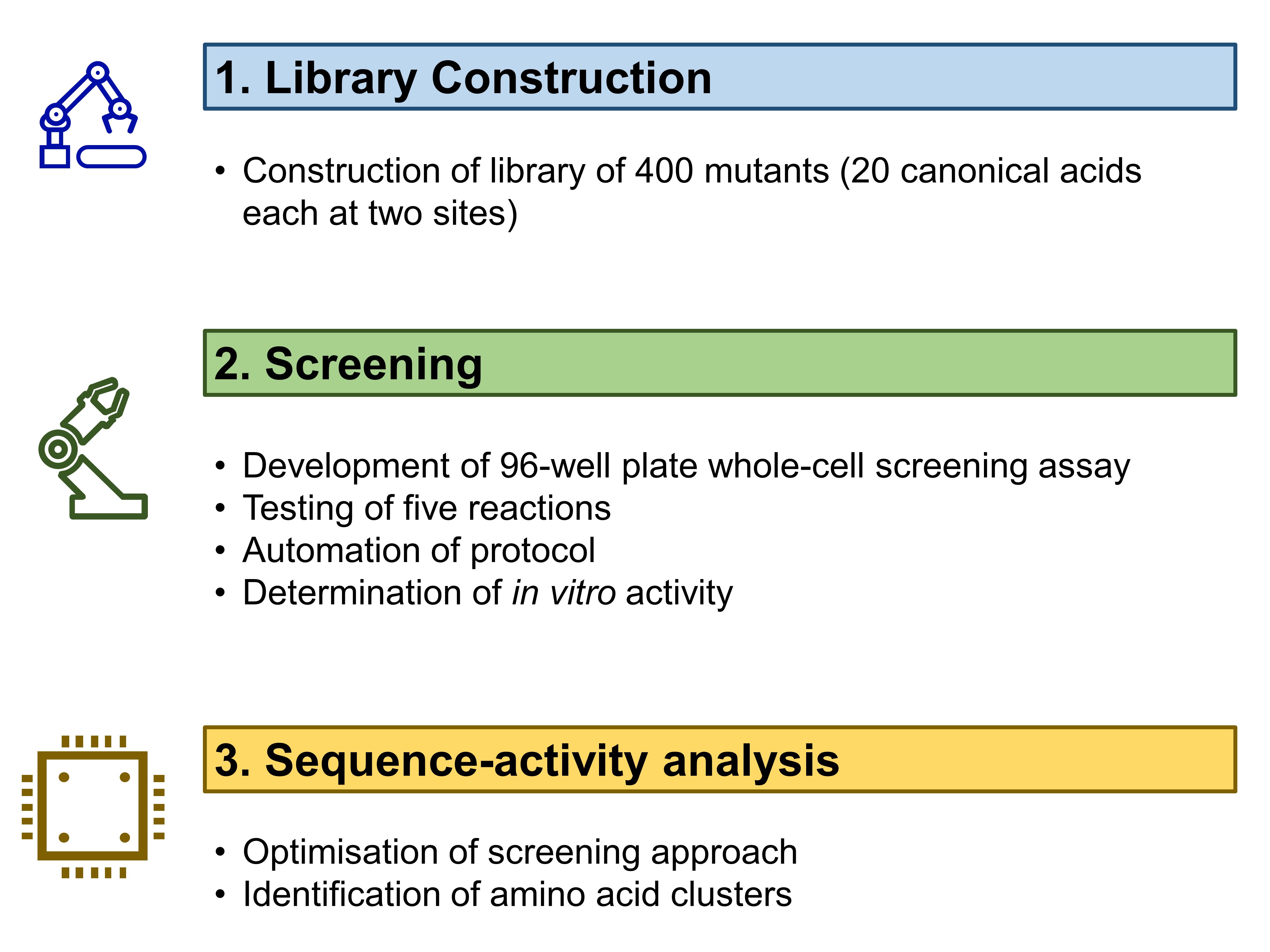
Figure 1. Work described by Vornholt et al. in their preprint.
(A) Construction of a sequence-defined Sav library
Building on the research group’s previous experiences with Sav expression and whole-cell ArM catalysis [1,2], the authors decided on a periplasmic compartmentalisation strategy for their library. Vornholt et al. chose to randomise amino acid residues 112 and 121 in Sav—residues that correspond to serine and lysine respectively in wildtype Sav—as mutation of these residues significantly affect the activity of ArMs [1-3]. By replacing the 112 and 121 positions each with the 20 canonical amino acids, the authors thus created a 20 x 20 = 400-mutant Sav library.
(B) Screening of ArMs for bioorthogonal reactions
Vornholt et al. then used a 96-well plate screening assay to screen the 400 mutants in their library. They selected five reactions to test for (Fig. 2): three of these were based on previously-reported ArMs, as well as hydroamination and hydroarylation. All five reactions are especially valuable given that none is catalysed by natural enzymes.
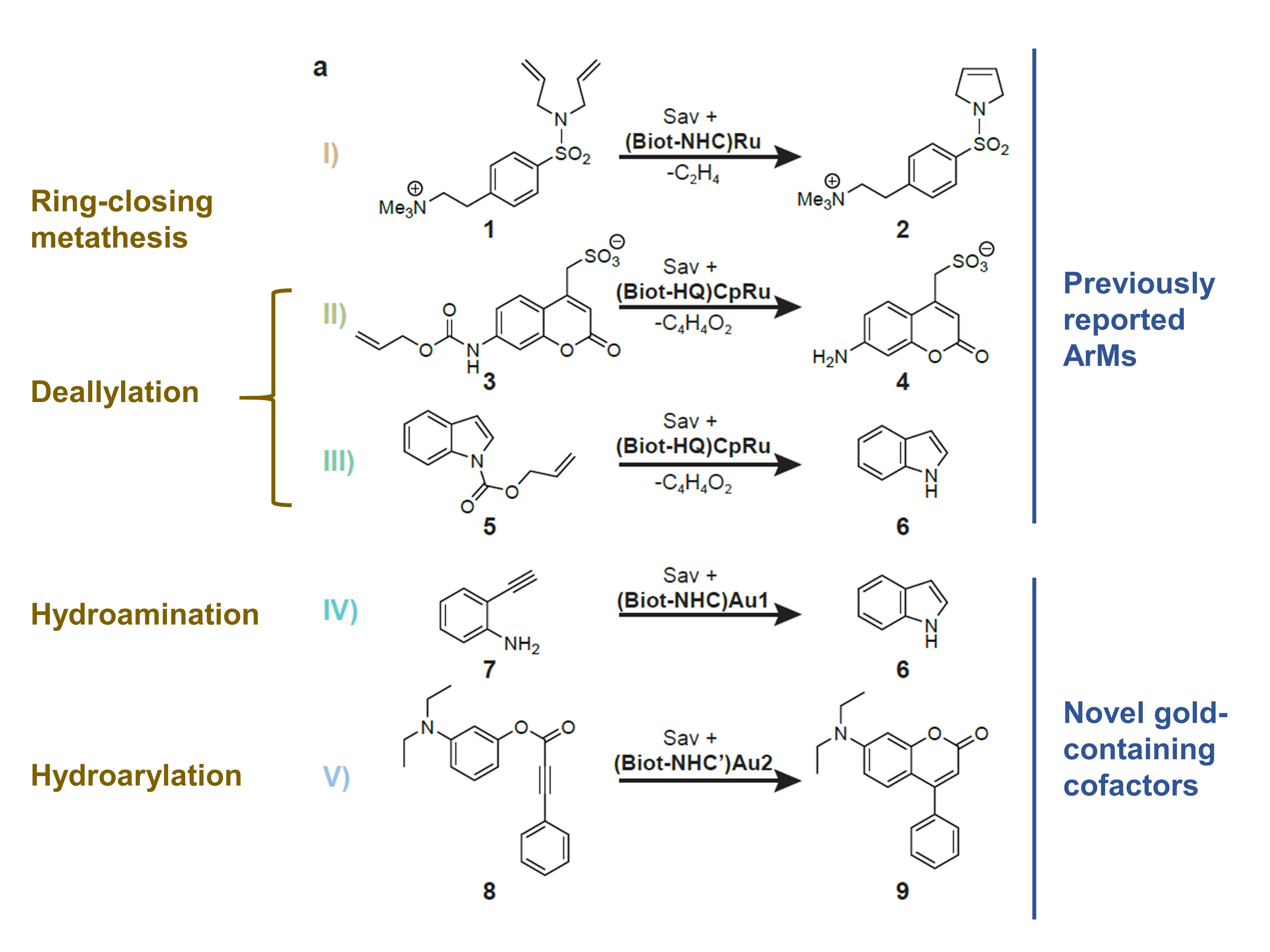
Figure 2. The five reactions investigated by Vornholt et al. in their preprint.
The authors found that reactions I, II, and III, but not IV and V, were compatible with their periplasmic screening platform. Further investigations revealed that this was due to the incompatibility of gold-catalysed reactions with cellular thiols, an effect that the authors mitigated by using the oxidising agent diamide.
Most importantly, the authors also automated the protocol for reactions II and V, a productivity-boosting step that allowed the authors to quickly and efficiently record a sequence-activity landscape for each ArM. These activity patterns varied substantially between reactions, leading them to conclude that there are specific interactions between protein, cofactor, and substrate (the authors also undertook further experiments ruling out unspecific interactions like varying expression levels).
Lastly, the authors also investigated the activity of selected ArMs in vitro, and found that the selected ArMs all outperformed the wildtype ArM as measured by turnover numbers (TONs).
(C) Analysis of sequence-activity landscapes
Having collected the data to generate sequence-activity landscapes for the ArMs, Vornholt et al. now analysed the effect of individual amino acids on the activity of the ArMs. The authors’ analysis revealed that amino acids with similar chemical properties—such as hydrophobicity and hydrophilicity—tended to cluster closely together.
Finally, Vornholt et al. then discussed their strategy of full-factorial screening, and compared it to other enzyme screening strategies like screening single mutants and iterative saturation mutagenesis. Because full-factorial screening would not be feasible as the diversity of mutants expanded, the authors discuss the need for alternative screening strategies. One such strategy would be to screen single mutants, but due to additive effects between mutations at positions 112 and 121, this would probably not have yielded the best combination. Another strategy, iterative saturation mutagenesis, would have considered these additive effects, but would be limited to the discovery of combinations that yield local optima, rather than global optima. To circumvent these problems, the authors propose that future work can involve screening with reduced amino acid sets in saturation mutagenesis: this strategy would have covered all the major clusters that they identified in their study.
What I like about this preprint
I selected this preprint because it highlights an important feature—automation—in the context of synthetic biology and bioengineering. Automation brings many benefits in addition to saving on human labour; it improves replicability (because it is a standardised process) and is more environmentally friendly. Many experiments involving biology are labour-intensive and can be thus be slow, labourious, and expensive; the single-use nature of many plastics in these experiments also contributes to a significant amount of waste.
Having a high-throughput, automated strategy that utilises 96-well plates therefore improves efficiency, allowing Vornholt et al. to screen many more reactions at once. Indeed, the authors themselves recognised that lab automation would, in tandem with their suggestion of using reduced amino acid sets, enable larger screening campaigns involving an expanded diversity of amino acid mutations.
Future work
With the formulation of a strategy amenable to automation by Vornholt et al., future efforts will consist of rolling this strategy out on two scales: research laboratories and academia will generally work on a small scale, while industrial companies tend to work on a larger one.
In research laboratories fortunate enough to be equipped with a robotic arm, screening efforts can be scaled up. Researchers may apply this strategy to ArMs with other metallocentres, such as copper, zinc, nickel, and iron. A more extensive in vitro study may also be undertaken, with ArMs optimised for their in vitro activity rather than their whole-cell activity.
Another possible research direction will also lie in answering questions related to the ArM-substrate specificity. Do ArMs work on substrates equally, or do they react with some substrates better than others? What gives rise to this selectivity? What conformations do the ArMs adopt during reactions, and what interactions form between the substrates and the binding site? The ability to introduce mutations on specified amino acid residues could allow researchers to optimise for these interactions and improve the specificity constant of an enzyme.
Finally, in moving these results to industry, a different set of questions emerges. How economically feasible is it to produce these ArMs, and once produced, how many reactions can they undergo before they degrade? Again, a massive screening effort must be undertaken to explore all the challenges associated with reusability, sustainability, and scale-up that such a campaign would involve.
All in all, this is a remarkable work by Vornholt et al. to advance the field of biocatalysis and synthetic biology. The implementation of such an automated strategy would benefit many stakeholders in multiple ways. It is a call to arms.
Open questions
- Why did you choose to automate reactions II and V?
- There are fewer complicating factors in vitro, in part because researchers can use chemically defined media, and exclude incompatible chemicals and cellular components. Would it be more feasible or desirable to shift towards in vitro work, or would a whole-cell approach be more viable?
- Why did you choose to work on Au and Ru? Did you consider working on ArMs with other metal centres, such as Co, Fe, and Zn?
References
[1] Heinisch T, Schwizer F, Garabedian B, Csibra E, Jeschek M, Vallapurackal J, Pinheiro VB, Marlière P, Panke S, Ward TR, E. coli surface display of streptavidin for directed evolution of an allylic deallylase, Chemical Science 9(24) (2018) 5383-5388.
[2] Jeschek M, Reuter R, Heinisch T, Trindler C, Klehr J, Panke S, Ward TR, Directed evolution of artificial metalloenzymes for in vivo metathesis, Nature 537(7622) (2016) 661-665.
[3] Hyster TK, Knörr L, Ward TR, Rovis T, Biotinylated Rh(III) complexes in engineered streptavidin for accelerated asymmetric C-H activation, Science 338(6106) (2012) 500-503.
doi: https://doi.org/10.1242/prelights.25271
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)