Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Posted on: 12 December 2025
Preprint posted on 20 September 2025
Categories: neuroscience, physiology
Background:
The foundational principle of physiology, articulated by Claude Bernard and later canonized by Walter Cannon, is that complex organisms maintain a stable internal milieu-homeostasis through a symphony of coordinated responses across organs1. This “wisdom of the body” relies on intricate feedback loops that span neural, endocrine, and immune systems, allowing an animal to adapt to external stressors and internal demands2. For decades, our understanding of these networks has been largely inferential, pieced together from studies that examine one organ, one pathway, or one molecule at a time. This approach, while invaluable, is akin to trying to understand an orchestral piece by listening to individual instruments in separate rooms; the score of the whole remains elusive. This gap between conceptual framework and observational capability has been a central challenge in biology and medicine3.
The rise of functional neuroscience offered a new paradigm for observing biological processes at scale. Pioneering work, particularly in transparent model organisms like larval zebrafish, demonstrated the power of using genetically encoded calcium indicators (GECIs) to monitor the activity of thousands of neurons simultaneously during behavior4,5. This “brain-wide” approach led to fundamental insights into sensorimotor processing6 and neural coding7, proving that observing a system in its entirety could reveal principles invisible to reductionist methods. However, this powerful lens remained almost exclusively focused on the central nervous system. The rest of the body, the very organs with which the brain continuously communicates to regulate physiology, was rendered a black box. This created a critical knowledge gap: we could decode a brain’s response to a threat, but not the concurrent, real-time cellular events in the immune system, gut, and vasculature that together constitute the full physiological response.
Bridging this gap presented monumental technical hurdles. Extending functional imaging to the entire body was not simply a matter of scaling up the field of view. The viscera exhibit complex, non-rigid motion that confounds standard image registration algorithms developed for the relatively stable brain8. Tissues outside the CNS have diverse optical properties that scatter light, degrading resolution, and no transgenic strategy existed for robust, non-toxic expression of sensors across the vast diversity of non-neuronal cell types. While recent studies provided glimpses into specific body-brain connections revealing, for instance, neurons that regulate inflammation9 or link gut cues to behavior10, these were still discovered and examined one circuit at a time. The field lacked a generalizable, unbiased platform to screen the entire vertebrate for coordinated cellular activity, a tool that could move beyond testing specific hypotheses to generating them de novo from a complete functional dataset.
In this groundbreaking preprint, Ruetten and colleagues directly confront this long-standing challenge. They present WHOLISTIC: a unified methodological platform that finally enables the comprehensive observation of cellular calcium dynamics across nearly all tissues of a living vertebrate. By converging key innovations, the authors achieve a systems-level view of physiology that has been a central goal of the field for decades. Furthermore, by integrating their functional imaging with a newly developed Whole-Body Expansion Microscopy (WB-ExM) protocol11,12, they provide the essential anatomical context, allowing them to trace the subcellular structures, such as the elaborate ventral projections of ependymal cells (Fig. 1), that underlie the observed body-wide dynamics.
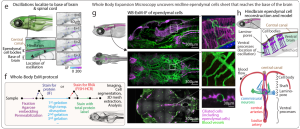
Fig. 1 – Propagation of Ca2+ waves and whole-body expansion microscopy of hindbrain ependymal cells. Fig. 1 of the preprint, made available under a CC-BY-NC-ND 4.0 International license.
Key Findings:
A unified platform for whole-body functional imaging (WHOLISTIC):
The authors engineered a Tg(ubi:tTA; TRE:GCaMP7f) zebrafish line for sustained, high-fidelity sensor expression across >15 cell types. Bypassing standard light-sheet microscopy, they identified spinning-disk confocal as uniquely capable for whole-body volumetric imaging due to its superior scattering rejection. Their custom iterative optical flow algorithm solves the critical problem of non-rigid motion in viscera, enabling stable tracking of individual cells over hours. The computational workflow segments individual cells and then uses a new coherence-based spectral clustering method to group them into functionally coupled anatomical units.
Discovery of New Functional Units and Couplings:
Applying WHOLISTIC as an unbiased discovery tool yielded multiple insights: a) Revised Muscle Anatomy: The preprint authors identified a novel functional synergy between the cervical-epaxial and abdominal muscles, linked to dual innervation by the spino-occipital nerve, revising classical anatomical maps. b) Body-Wide Motor Coupling:A regression model revealed that skeletal muscle activity triggers calcium transients in diverse tissues. A key discovery was that ependymal cells, glia-like cells lining the central canal, show prolonged, movement-locked calcium signals. c) State-Dependent Ultraslow Oscillations: During extended motor quiescence, a network of hindbrain/spinal cord ependymal cells exhibited high-amplitude, traveling calcium waves with a ~3-7 minute period, potentially linked to rest-state physiology. d) Causal Vagus-Sphincter Circuit: Combining WHOLISTIC with optogenetics, the research team validated a direct, topographical circuit from motor vagal neurons to the gastropharyngeal sphincter, demonstrating causal control of visceral smooth muscle.
Visualizing a Conserved Hypoxia Response Circuit:
Under hypoxic conditions, WHOLISTIC captured the rapid constriction of the mesenteric artery and shunting of blood flow away from the gut in real-time, a classic stress response never before imaged dynamically at cellular resolution. The researchers proved this is neurally regulated: optogenetic inhibition of the hindbrain instantly restored gut perfusion. WB-ExM then revealed the responsible sympathetic ganglion and its axonal projections along the artery, defining a complete brain-body circuit for oxygen prioritization (Fig. 2).
Anatomical Ground Truth with WB-ExM:
The team developed an enzyme-free, rapid WB-ExM protocol for ~5x uniform expansion of whole zebrafish and Danionella, compatible with immunofluorescence and in situhybridization. This provided the essential ultrastructural context for their functional observations, enabling high-resolution molecular phenotyping across the entire organism.
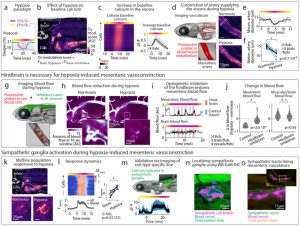
Fig. 2 – Uncovering of body-wide circuit engaged in response to stress. Fig. 2 of the preprint, made available under a CC-BY-NC-ND 4.0 International license.
Why I highlight this preprint?
This preprint represents the kind of transformative science that redefines what is possible in a field. As someone working in zebrafish systems neuroscience, I have admired the Ahrens lab’s philosophy: to build the tools needed to answer the biggest, most integrative questions. Misha has consistently pushed the boundaries of in vivo imaging, and WHOLISTIC feels like the culmination of this vision: a platform that shifts the very unit of observation from the brain to the entire organism. It’s a breathtaking technical and conceptual achievement that embodies the collaborative, tool-building spirit of Janelia Farm.
My excitement for this work is multifaceted. It solves a fundamental measurement problem. For decades, we’ve studied physiology in fragments. WHOLISTIC provides the first unified lens to see the organism as an integrated circuit, where the state of a kidney tubule, a skin cell, and a neuron can be correlated simultaneously. This is not merely incremental; it’s a paradigm shift from reductionism to holistic systems biology, echoing the foundational principles of homeostasis but now with the tools to visualize it.
The methodology itself is a masterpiece of integration. Each component from the pancellular driver and motion correction to the coherence-based clustering and WB-ExM is a significant innovation. Together, they form a closed loop of discovery: unbiased functional imaging generates hypotheses, optogenetics tests causality, and expansion microscopy provides mechanistic anatomical explanation. This pipeline sets a new standard for in vivo physiology.
Finally, the implications are vast. Beyond the immediate, fascinating discoveries about muscle synergy, ependymal networks, and hypoxia circuits, WHOLISTIC opens entirely new frontiers. For disease modelling and drug discovery, it offers a platform to screen for organism-wide effects and off-target actions, moving beyond single-organ endpoints. For basic research, it enables the study of previously intractable problems like inter-organ communication during sleep, stress, or metabolic regulation. This work doesn’t just advance zebrafish research; it provides a blueprint for holistic biology that could eventually inform a new understanding of human physiology and integrative medicine. It is, in every sense, a foundational study for the next era of systems biology.
Reference:
- Cannon, W. B. (1939). The wisdom of the body.
- Billman, G. E. (2020). Homeostasis: the underappreciated and far too often ignored central organizing principle of physiology. Frontiers in physiology, 11, 200.
- Buchman, T. G. (2002). The community of the self. Nature, 420(6912), 246-251.
- Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M., & Keller, P. J. (2013). Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nature methods, 10(5), 413-420
- Vladimirov, N., Mu, Y., Kawashima, T., Bennett, D. V., Yang, C. T., Looger, L. L., … & Ahrens, M. B. (2014). Light-sheet functional imaging in fictively behaving zebrafish. Nature methods, 11(9), 883-884.
- Naumann, E. A., Fitzgerald, J. E., Dunn, T. W., Rihel, J., Sompolinsky, H., & Engert, F. (2016). From whole-brain data to functional circuit models: the zebrafish optomotor response. Cell, 167(4), 947-960.
- Portugues, R., Feierstein, C. E., Engert, F., & Orger, M. B. (2014). Whole-brain activity maps reveal stereotyped, distributed networks for visuomotor behavior. Neuron, 81(6), 1328-1343.
- Voleti, V., Patel, K. B., Li, W., Perez Campos, C., Bharadwaj, S., Yu, H., … & Hillman, E. M. (2019). Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nature methods, 16(10), 1054-1062.
- Jin, H., Li, M., Jeong, E., Castro-Martinez, F., & Zuker, C. S. (2024). A body–brain circuit that regulates body inflammatory responses. Nature, 630(8017), 695-703.
- Tan, H. E., Sisti, A. C., Jin, H., Vignovich, M., Villavicencio, M., Tsang, K. S., … & Zuker, C. S. (2020). The gut–brain axis mediates sugar preference. Nature, 580(7804), 511-516.
- Chen, F., Tillberg, P. W., & Boyden, E. S. (2015). Expansion microscopy. Science, 347(6221), 543-548.
- Ruetten, V. M., Hu, A., Eddison, M., Close, K., He, Y., Ahrens, M. B., & Tillberg, P. (2025). WHOLISTIC ExM: Whole-Body Expansion Microscopy with Immunofluorescence and Histological Stains.
doi: https://doi.org/10.1242/prelights.42477
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the neuroscience category:
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
Also in the physiology category:
Trade-offs between surviving and thriving: A careful balance of physiological limitations and reproductive effort under thermal stress
Tshepiso Majelantle
Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Muhammed Sinan Malik
Wide-ranging behavioral dysfunction in two mouse models of pathological human variants in the GRIK2 kainate receptor gene
Pushpinder Singh
preLists in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Also in the physiology category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |











 (No Ratings Yet)
(No Ratings Yet)