A spatial map of antennal-expressed olfactory ionotropic receptors in the malaria mosquito
Posted on: 2 June 2022 , updated on: 11 May 2023
Preprint posted on 12 May 2022
Would an olfactory receptor in any other place smell just as sweet? Ionotropic receptor position on the mosquito antenna.
Selected by T. W. SchwanitzCategories: bioinformatics, molecular biology, neuroscience
Updated 11 May 2023 with a postLight by T. W. Schwanitz
This study was published in Cell Reports with numerous intriguing updates and revisions. A third author is now on the paper, underscoring just how much got added. The most fascinating updates for me are the functional and behavioral follow-ups. The original preprint relied on in situ probes, so the inclusion of supporting data from a transgenic line for IR41c that almost perfectly matches the conclusions from the in situ probes is very reassuring. The calcium imaging results for these IR41c neurons reveal unexpected inhibitory responses to some odorants.
The authors addressed one initial question by including additional age groups in their study. The different age groups didn’t show any changes in in situ IR gene expression, which means that differences in the number of IRs expressed likely do not result from gross changes in the number of IR neurons. Instead, these changes in expression could be the result of an increase in the number of IR receptors per neuron.
The heatmap in what is now figure 1E has changed: the authors kept the version organized by transcript abundance and altered the color scheme. Overall, the new color scheme is nicer, but it does obscure some of the stand-out IRs a bit, instead putting emphasis on the co-receptors. IR41c, for example, is a bit of an outlier in a few of the studies, but that is not very obvious with the new color scheme. This is an instance where consulting both the preprint and the final paper can assist with data visualization.
Overall, this is a fascinating and detailed study where a lot was added between the preprint and the final publication: numerous additional figures and panels help to portray the results, and substantial textual revisions help the reader to contextualize the findings.
Background: Location, location, location: as with real estate, location matters for olfactory receptors. Mosquito olfactory receptors are mostly located on the antennae (Fig. 1A). But how are olfactory receptors distributed there, and how does that affect their sense of smell? In Drosophila melanogaster, the structures that house olfactory receptors—the sensilla—are localized to specific regions of the antennae (Vosshall and Stocker, 2007). In contrast, mosquito antennae have a more regular distribution of sensilla (Seenivasagan et al., 2009). Nevertheless, when Raji and Potter investigated the spatial distribution of the ionotropic receptors housed within these sensilla, they found that the receptors were not evenly distributed.
To detect airborne molecules, the mosquito olfactory system uses many different types of receptors, such as gustatory receptors, odorant receptors, and ionotropic receptors. The latter are ion channels that have a binding site for specific odorants. Usually, a “tuning ionotropic receptor” is tuned to a certain odorant or class of odorants. It forms a complex with an “ionotropic coreceptor,” an ionotropic receptor that isn’t linked to just one group of compounds. Ionotropic receptors appear to be especially good at detecting acids and amines; hence, they may influence the preference some mosquitoes have for biting human beings, as amines could be one component of human odor that attracts mosquitoes (Chen et al., 2019).
The two species investigated in this preprint are both members of the confusing Anopheles gambiae species complex: An. gambiae s. s. (S form) and An. coluzzii (M form). While the exact taxonomic classification of these mosquitoes is unclear, their effect on humanity is hard to overlook—the malaria parasite that they transmit has been and still is one of the deadliest human pathogens. They make such good vectors because of their predilection for biting human beings and their preference for human odor. Deeper understanding of mosquito ionotropic receptors could facilitate better mosquito control methods.

Fig. 1A. Head of the Anopheles gambiae mosquito, with a schematic to show the antennae and mouthparts.
Key findings:
- Using whole-mount fluorescent in situ hybridization for four tuning ionotropic receptors (IRs 7w, 41c, 41t.2, 75l) and three ionotropic coreceptors (IRs 25a, 76b, and 8a), the authors show that these receptors are not evenly distributed on the antenna. Generally, tuning ionotropic receptors tend to be on the part of the antenna furthest from the head. Even two of the three coreceptors tested (IRs 25a and 76b) were most expressed on the distal half of the antenna (while IR8a seems to be common throughout the antenna; see Fig. 2).
- These results were consistent across multiple individuals, though there are notable differences between males and females, e.g., males do not express IRs on the first ten segments closest to their head and IR41t.2 expression is enriched in male antennae.
- The authors conducted a transcriptome analysis and then used that data to conduct in situ probes of all 20 additional ionotropic receptors that they found upregulated in the transcriptome. Again, they revealed that these receptors tend to be most expressed toward the distal end of the antenna. IR93a makes for a striking example, as 80% of the cells expressing it were on the 13th flagellomere, which is the final antennal segment at the very tip. Testing a limited number of these receptors in males confirmed the trend observed in females.
- Next, Raji and Potter used two probe whole mount in situs to examine co-expression among ionotropic coreceptors and four highly expressed tuning receptors. They found numerous interesting trends, e.g., IRs 7w and 75l overlap with IR8a, a coreceptor that has been previously linked to acid-sensitive tuning ionotropic receptors. This overlap suggests that the two tuning receptors could also be linked to acids. These results show that simply examining patterns of intersection can lead to hypotheses about receptor function.
- The authors also found that the number of ionotropic receptors expressed per flagellomere (antennal segment) seems to be consistent across individuals; however, the exact location of a receptor within a flagellomere is highly variable among individuals.
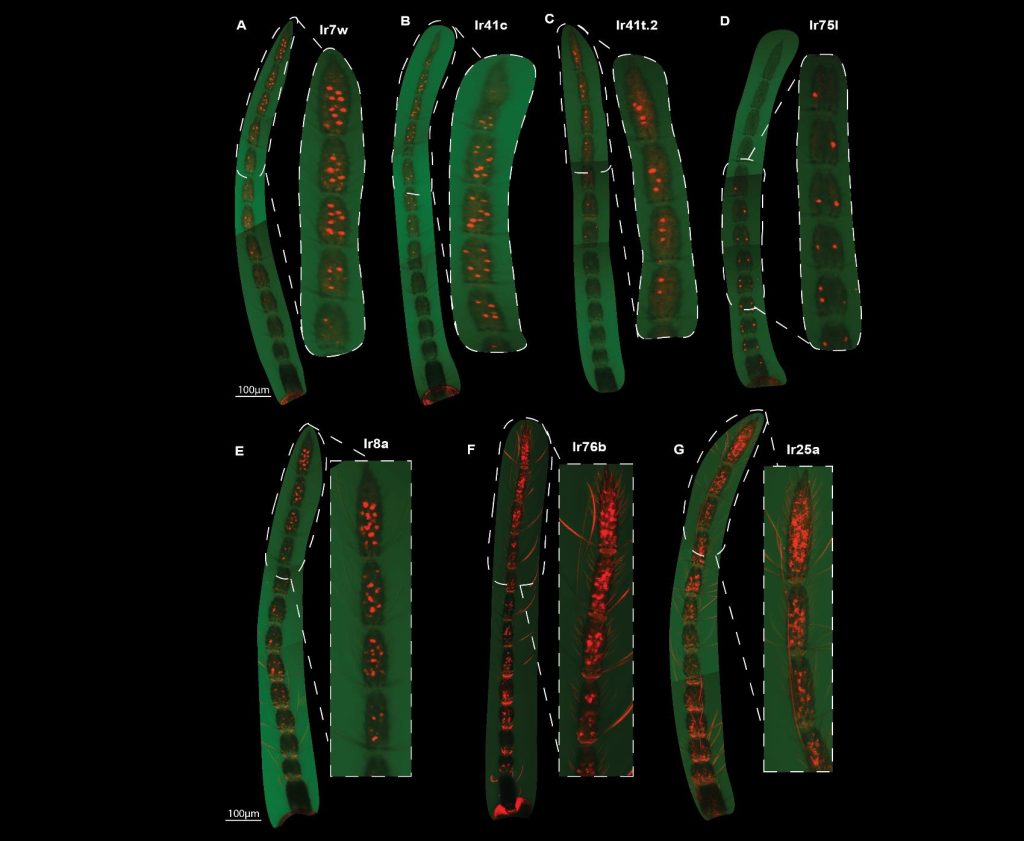
Fig. 2. Different receptors fluorescing along the mosquito antennae. Next to each full antenna is an inset of the region with the highest expression. Note that in D, IR751 is most expressed toward the proximal part of the antenna while the other receptors shown are most expressed in the distal portion.
Questions for the authors:
- You mention that your study is a “snapshot” of the expression of ionotropic receptors in mosquitoes that are five-to-seven days old. Are there any plans to look at other age ranges? Could there be any interesting insights from looking at ionotropic receptor expression in larvae or pupae? And would your in situ probes be possible in larval antennae?
- There are a lot of tantalizing instances of co-expression in your paper that seem like they could be worth follow-up studies, e.g., IR751 and IR8a being co-expressed suggests that IR751 is likely sensitive to acids. If you could only do functional studies on one of these combinations (empty neuron studies, for example), which combination would you investigate further, and why?
- Any speculation as to why ionotropic receptors are most expressed on the distal portion of the antenna and not the proximal portion?
- Do you think that odorant receptors could share a similar pattern of distribution along the antenna?
Why I think this study is important:
Where is it? What is it? How does it work? These are some of the most fundamental questions that can be asked about something, and they serve as the basis for more complex follow-up questions. By shedding light (and fluorescence) on the location of ionotropic receptors, Raji and Potter lay the groundwork for more precise studies. Knowing that a given receptor is mostly found on the distal part of the antenna means that studies investigating this receptor could focus on collecting only that part of the antennae, thus reducing background noise. Moreover, the authors make use of an interesting method that can reveal new insights about the function of different receptors.
References:
Chen, Z., Liu, F., & Liu, N. (2019). Human odour coding in the yellow fever mosquito, Aedes aegypti. Scientific reports, 9(1), 1-12.
Seenivasagan, T., Sharma, K. R., Shrivastava, A., Parashar, B. D., Pant, S. C., & Prakash, S. (2009). Surface morphology and morphometric analysis of sensilla of Asian tiger mosquito, Aedes albopictus (Skuse): an SEM investigation. Journal of vector borne diseases, 46(2), 125.
Vosshall, L. B., & Stocker, R. F. (2007). Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci., 30, 505-533.
doi: https://doi.org/10.1242/prelights.32219
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the molecular biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Also in the neuroscience category:
Electrophysiological correlates of conscious experiences during sleep: Lucid dreams, sleep paralysis, out-of-body experiences, and false awakenings
uMontreal Neuro preLighters et al.
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)