COMET: A toolkit for composing customizable genetic programs in mammalian cells
Posted on: 29 September 2019
Preprint posted on 16 September 2019
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-019-14147-5
Reaching for the shooting stars: a COMET toolkit for bespoke mammalian cell genetic programming
Selected by Zhang-He GohCategories: bioengineering, synthetic biology
Background of preprint
Prokaryotes are commonly regarded as model systems in the field of cellular biology [1] and used to produce useful small molecules and proteins in the fields of theranostics [2,3]. In contrast, owing to limitations on the transfection of eukaryotes, mammalian cells remain relatively intractable despite Herculean efforts. In their preprint, Donahue et al. suggest that three factors are needed to establish a transcriptional toolkit for cell engineering (Figure 1).
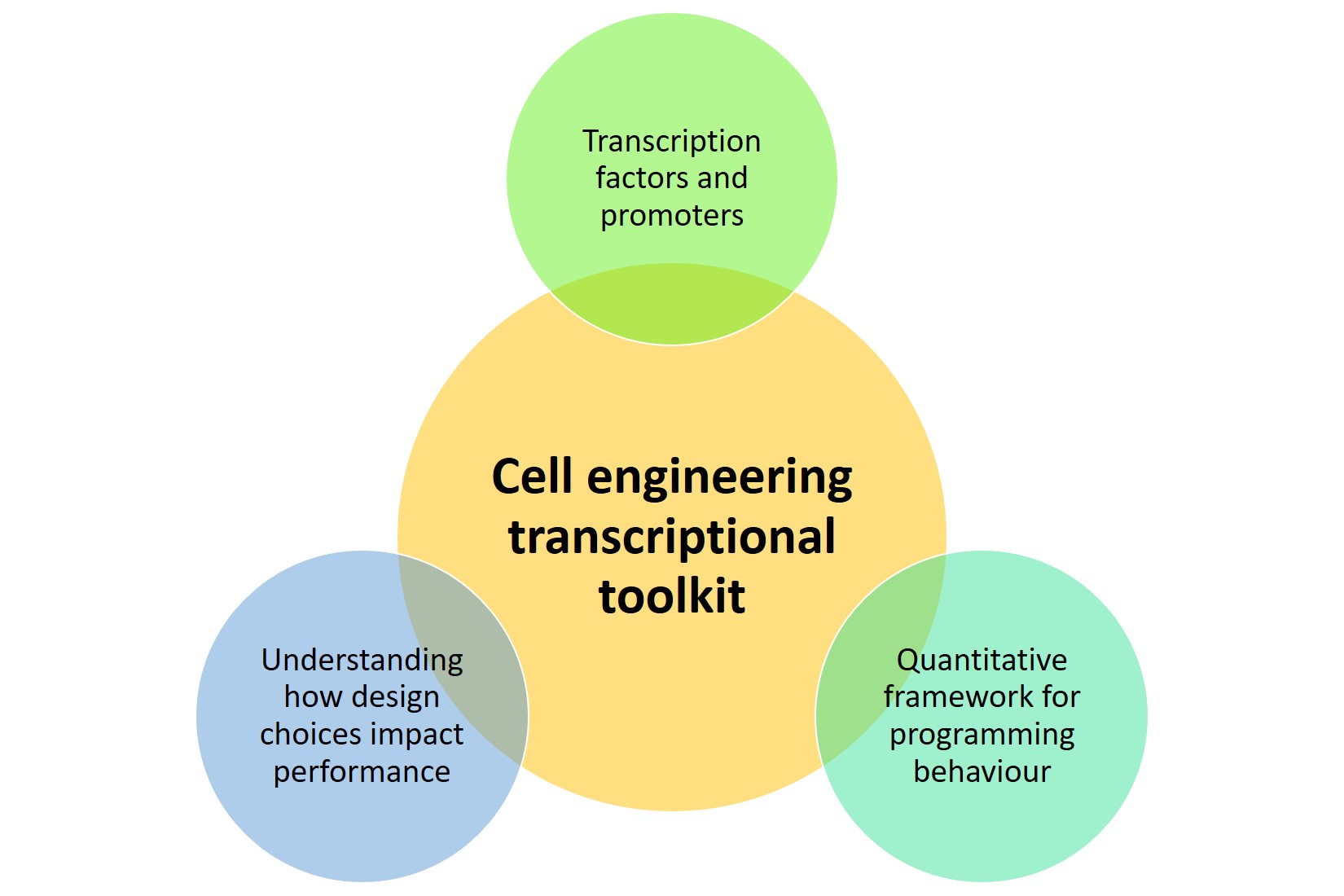
Figure 1. Factors considered in establishing a transcriptional toolkit
To answer their research question, Donahue et al. developed and characterised the COmposable Mammalian Elements of Transcription (COMET), described as “an ensemble of engineered promoters and modular zinc finger-transcriptional factors with tunable properties”. The authors then used their toolkit to (a) elucidate design rules to incorporate activation, inhibition, small molecule responsiveness, and Boolean logic into mammalian cells, (b) develop a mathematical model to describe these respective properties, and (c) characterise new promoters and modified zinc fingers.
Key findings of preprint
(A) Promoter design rules
Donahue et al. first examined five synthetic zinc finger domains that were originally characterised in yeast [4] and adapted them to regulate transcription in a human human embryonal kidney cell line (HEK293FT). From this optimisation process, the authors concluded that three variables—the number, placement, and compaction of binding sites—could be tuned to modulate transcriptional output.
(B) Mathematical model
Donahue et al. observed that zinc finger activator-induced gene expression is transcriptionally, rather than translationally inhibited. This led the authors to use a mathematical model to describe the trends that they observed. While the intent of designing this model was not to predict novel transcriptional factor or promoter sequences, Donahue et al. believe that this framework could be used to extend the capabilities of their COMET platform.
Specifically, Donahue et al. observed that maximum transcription rate increases with both the number and compactness of binding sites; however, when relative reporter output was maximised, two trends were observed (preprint Supp. Fig. 3d):
- For each binding site series (at 3x, 6x, and 12x), the dependence of relative reporter output on the number of binding sites was independent of the dose of zinc finger activator plasmid.
- For each dose series (ranging from 5 – 200 ng), the dependence of relative reporter output on zinc finger activator dose was independent of the number of binding sites.
(C) New promoters and modified zinc fingers
Donahue et al. then tested whether the established design rules could be applied to the design of other zinc finger activators. The authors first showed that zinc finger activators are orthogonal (preprint Fig. 3c): among the 144 pairs of zinc finger activator and promoter tested, 12 were between zinc finger activator and cognate promoter. This left 132 pairs of zinc finger activator and non-cognate promoter; of these, 80% showed less than 1% off-target activation, and 97% showed less than 3% off-target activation.
The preprint authors then showed that gene expression could also be modulated through transcription factor engineering, specifically by altering (a) the affinity of zinc fingers for DNA, and (b) the strength of the activation domain. Taken together, these results led the authors to identify physical features that can be optimised to modulate gene expression (Figure 2).
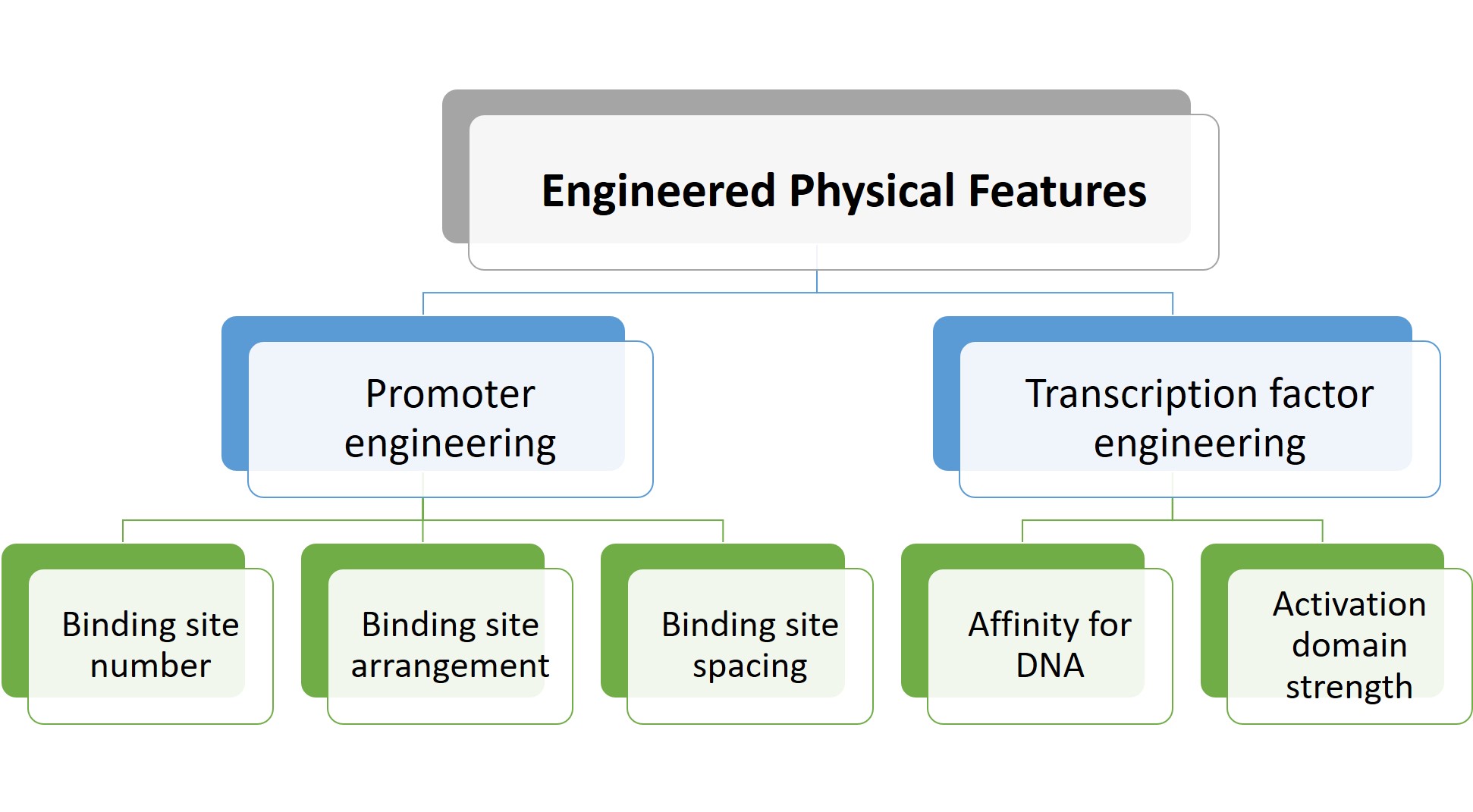
Figure 2. Adjustable physical features to modulate gene expression
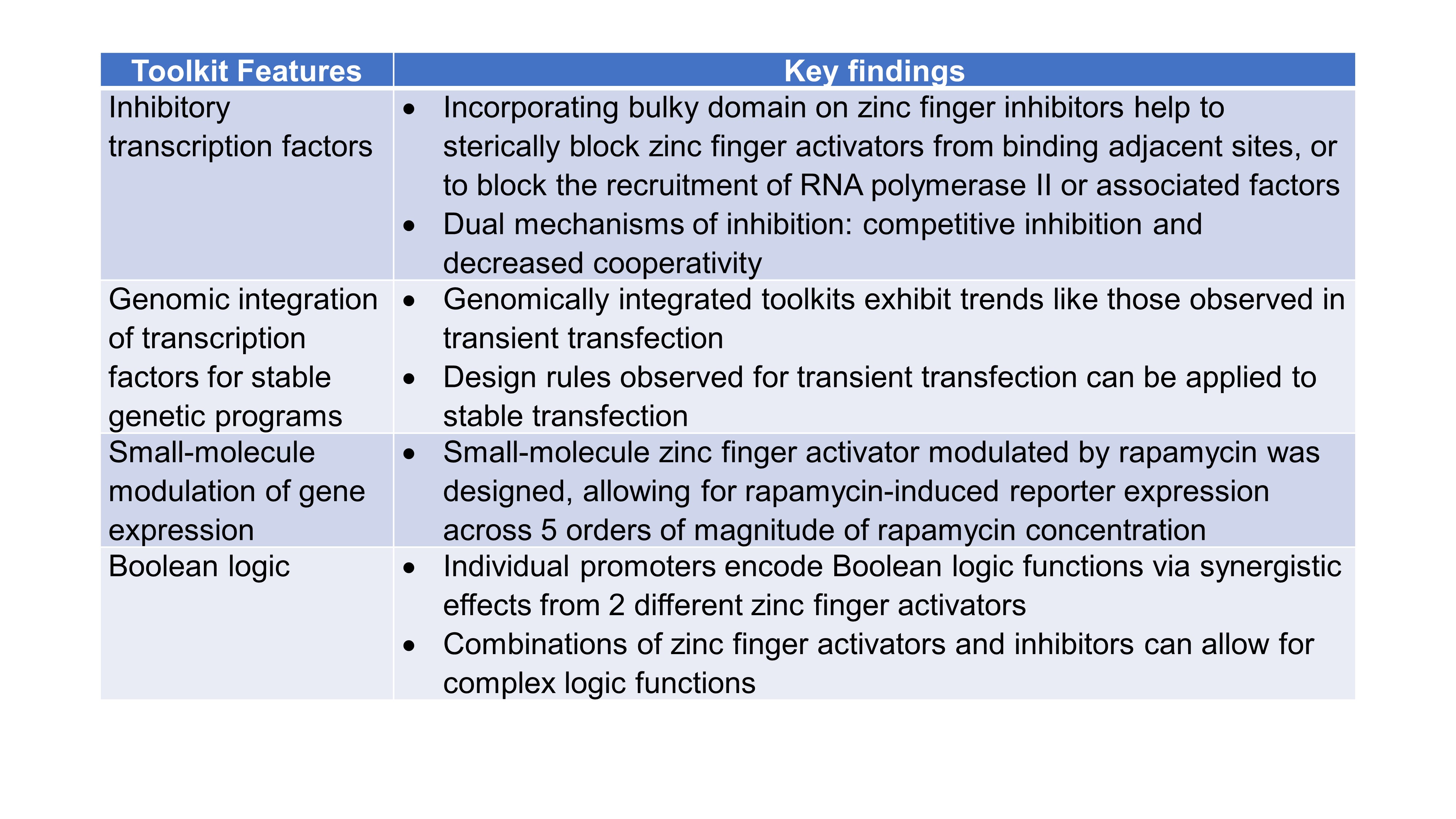
Finally, Donahue et al. considered potential applications and designed four additional features that could be used in their toolkit (Table 1): (i) inhibitory transcription factors, (ii) genomic integration of transcription factors for stable genetic programs, (iii) modulation of gene expression using small molecules, and (iv) implementation of Boolean logic.
Table 1. Summary of COMET Toolkit Features
What I like about this preprint
By demonstrating a proof of principle and describing a framework by which their system can be optimised, this preprint by Donahue et al. addresses a growing need to make mammalian cellular systems easier to manipulate. Perhaps most directly, this most benefits the twin fields of pharmacology and toxicology, which has started adopting the 3Rs of animal research—replacement, reduction, and refinement. In fact, just earlier this week, the US Environmental Protection Agency announced a shift away from using animals in safety and toxicity studies. Indeed, researchers already started turning away from in vivo animal models, instead opting to conduct their research using in vitro and in silico models that stretch their research dollars and reduce animal suffering.
The enhanced tractability of mammalian cells will give other biology fields a fillip as well. Novel small molecules and proteins can be designed to aid in the elucidation of molecular pathways, making their mechanisms easier to interrogate, and possibly even giving rise to new molecular targets in drug discovery. Characterisation of these pathways, too, will become simpler, and careful design of rescue experiments will help to validate potential molecular targets.
The simple modification of mammalian cells will also help in the production of biologics. Currently, multifactorial considerations need to be made in bioengineering and bioprocessing—ranging from the choice of the cell line to the optimisation of growth conditions. The preprint authors have compartmentalised this process into a few key variables within a valuable toolkit and framework, which may help industry players develop optimised cell lines more quickly and cheaply.
All in all, Donahue et al. describe a powerful approach to rethink and redesign cellular engineering. Some work remains before the COMET toolkit’s potential in synthetic biology will be fully realised: as the preprint authors point out, a priority would be to expand the toolkit to include additional transcriptional factors and cognate promoters for which they are selective. But, to channel Disney’s Hercules, the utilisation of this toolkit is not in a far off place. Watch it go the distance.
Questions for authors
- In addition to the preferential binding of zinc finger activators to their cognate promoters, do the two key trends observed in Supplementary Figure 3d also support the orthogonal nature of the variables you used in the model? While designing the mathematical model, did you observe other variables that were not fully independent, in that they may be correlated with each other in some way?
- You mentioned that “the observed trends will extend across cell types and delivery methods”. Indeed, it has been reported that transcription factors are overexpressed in certain cancers [5], so it is reasonable to postulate that these transcription factors may also be expressed in cell lines derived from these disease states. Considering these observations, what challenges do you foresee in translating these findings to these other mammalian cell lines, whether human or non-human? How might these difficulties be overcome?
References
[1] Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA, Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems, Annu Rev Biophys 39 (2010) 515-537.
[2] Muldoon JJ, Donahue PS, Dolberg TB, Leonard JN, Building with intent: technologies and principles for engineering mammalian cell-based therapies to sense and respond, Curr Opin Biomed Eng 4 (2017) 127-133.
[3] Lim WA, Designing customized cell signalling circuits, Nat Rev Mol Cell Biol 11(6) (2010) 393-403.
[4] Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ, A synthetic biology framework for programming eukaryotic transcription functions, Cell 150(3) (2012) 647-658.
[5] Lambert M, Jambon S, Depauw S, David-Cordonnier M-H, Targeting Transcription Factors for Cancer Treatment, Molecules (Basel, Switzerland) 23(6) (2018) 1479.
doi: https://doi.org/10.1242/prelights.14175
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)