Conditional Guide RNAs: Programmable Conditional Regulation of CRISPR/Cas Function in Bacteria via Dynamic RNA Nanotechnology
Posted on: 18 February 2019
Preprint posted on 21 January 2019
Article now published in ACS Central Science at http://dx.doi.org/10.1021/acscentsci.9b00340
gRNAs for Cas9 are engineered such that they only get activated upon specific RNA triggers. This could one day open the door to linking signaling pathways with Cas9 function!
Selected by Pavithran RavindranCategories: bioengineering
Background
The ability to build synthetic signaling circuits and modulate natural biological pathways is a goal of modern synthetic biology. Dynamic RNA nanotechnology is a very promising candidate to be able to accomplish this, especially for pathways that recognize RNA for initiation such as RNA interference. A platform that seems ripe for engineering is gRNAs for CRISPR/Cas9. There has already been a lot of interesting work done in this field to expand the uses of gRNAs and Cas9 to different functions other than the typical editing. For instance, work has been done to engineer the Cas9 protein itself such that it causes up-regulation or down-regulation of genes [1] or, more recently, target specific sites of the genome to particular locations in the cell [2]. gRNAs also provide a flexible platform for such engineering as researchers have introduced protein binding RNA motifs (MS2, PP7, etc.) into gRNAs to recruit protein effectors that can regulate genes of interest [3]. However, work still needs to be done to be able to spatio-temporally restrict these activities. The authors of this preprint set out to engineer conditional gRNAs that only perform their given function upon binding or release from particular RNA trigger sequences.
Key Findings
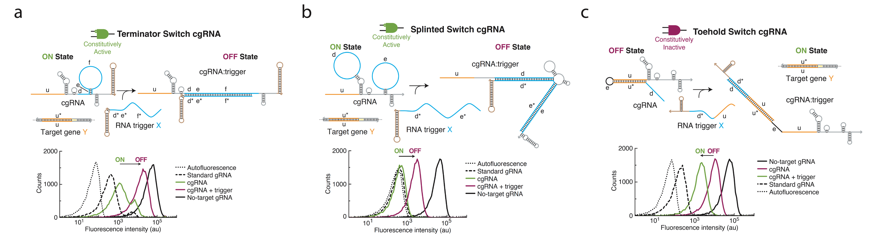
The authors set out to create conditional gRNAs (cgRNAs) that only interface with Cas9 upon binding to or unbinding from a particular RNA trigger sequence. By looking at the structure of a typical gRNA, the authors realized that the Cas9 handle sequence as well as the S. pyrogenes terminator sequence could be engineered to bind/unbind RNA trigger sequences to allow for interfacing with Cas9 and thus various effector functions described above (figure 1). To test the cgRNAs that they developed, the authors utilized E. coli expressing a reporter fluorescent protein, and catalytically inactive Cas9 (dCas9) that silences expression of the fluorophore. Therefore, when the cgRNA is ON, then the dCas9 would bind to the gRNA and the reporter gene, resulting in low fluorophore expression – whereas when the cgRNA is OFF then the fluorophore is not silenced. As controls to see what ON and OFF could look like, the authors introduced non-engineered gRNAs (for ON function) and modified gRNAs without the target-binding domain (for OFF function).
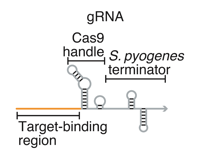
As a first pass at building cgRNAs, the authors decided to build one that operates in an ON to OFF logic fashion; thus, when the RNA trigger is introduced, the trigger binds to the cgRNA and inactivates it by preventing it from interfacing with dCas9 and thereby results in increased mRFP expression. The authors first tried to introduce a sequence into the terminator sequence of the gRNA that binds to a specific RNA trigger sequence. Binding would linearize this portion of the cgRNA, effectively turning it off . Upon adding the RNA trigger to the cgRNAs, it was very clear that mRFP expression was much higher compared to the case where the cgRNAs were in their ON conformation. However, the authors very quickly realized that the ON state of this cgRNA was not perfect as mRFP expression was higher than what was seen in the standard gRNA case, implying that the Cas9 was not as efficient at binding this engineered gRNA. Therefore, the authors tried to engineer the Cas9 handle and the terminator simultaneously. With this system the authors utilized sfGFP as their readout. The authors found that the ON conformation was very close to that of the standard non-engineered gRNA but this also resulted in a less optimal OFF conformation where, after the trigger was added, the cgRNA still seems to have some silencing ability. The beauty of this particular system is that, since the engineering occurred only in the constant regions of the gRNA and not the targeting sequence portion, this conditional activation of the gRNAs can be done independent of what gene the gRNA was meant to target.
The authors then tried to create a cgRNA that goes from an OFF state to an ON one. To achieve this, they added an extra sequence on the 5’ end of the gRNA that wraps around and covers up the targeting sequence. When the trigger is introduced, the trigger RNA binds to the part of the cgRNA covering the targeting sequence, effectively revealing the targeting sequence and making the cgRNA functional. This ON cgRNA was then shown to interface with dCas9 to silence mRFP expression (Figure 2C).
Why I chose this preprint
CRISPR/Cas9 has a very impressive suite of effector functions not limited to the original editing function that was first conceptualized. However, it is tough to limit these functions to particular cell types or during a particular phase of, say, development. This work begins to flirt with this level of control as they show that the cgRNAs can be modulated with the introduction of certain trigger RNAs. This opens the door to linking gRNA/Cas9 effects to signaling pathways in cells such that one could imagine activating Cas9 only in certain tissues within the body or after certain RNAs are expressed during development. This preprint was overall a fun and interesting read with figures that clearly show the results of the experiments.
Questions for the authors
Does changing gRNA structure impair double strand cutting ability of Cas9?
When creating your cgRNAs for ON to OFF, you altered the terminator sequence or the terminator and the Cas9 handle. Did you try the Cas9 handle alone and, if so, how did that work?
The utilization of NUPACK to find pairs of cgRNAs and trigger RNAs was used in this study. Can NUPACK or some other program be used to find trigger RNAs that are downstream/upstream of pathways such as Toll-like receptor (TLR), Rig-1 or RNA interference such that the cgRNAs can be activated with particular viral RNAs, mRNAs of signaling pathway targets, etc?
References
[1] Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173-83. PMID: 23452860.
[2] Wang H, Xu X, Nguyen CM, et al. CRISPR-Mediated Programmable 3D Genome Positioning and Nuclear Organization. Cell. 2018;175(5):1405-1417.e14. PMID: 30318144.
[3] Zalatan JG, Lee ME, Almeida R, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160(1-2):339-50. PMID: 25533786.
doi: https://doi.org/10.1242/prelights.8500
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |











 (No Ratings Yet)
(No Ratings Yet)