Glutamate helps unmask the differences in driving forces for phase separation versus clustering of FET family proteins in sub-saturated solutions
Posted on: 3 October 2023 , updated on: 5 October 2023
Preprint posted on 13 August 2023
Kar, et al. examine the effect of different ions on the behavior of a self-associative protein to better understand organization within the cell. This work highlights the exquisite mechanisms that exist to organize the complex cellular environment.
Selected by Erich SohnCategories: biochemistry, biophysics
Background
The internal cellular environment is organized through membrane-bound and membrane-less organelles (MLOs). MLOs are groupings of macromolecules that are not separated from the rest of the cell by any physical barrier [1]. The underlying interactions between these heterotypic macromolecules are the driving force for the formation and selectivity of MLOs [1]. Investigating how these interactions occur and are affected by the composition of the local cellular milieu helps us understand how the cell organizes itself functionally.
The self-associative properties of intrinsically disordered proteins (IDPs) are often central to the formation of MLOs [2]. This self-association is even seen in sub-saturated solutions, where visible de-mixing of the solution does not occur, and the interactions only result in transiently associating complexes of varying size [3]. In vitro, we often observe these interactions in super-saturated conditions, wherein molecules de-mix from the solution, akin to an oil-in-water emulsion, with droplets forming and fusing with one another. This phenomenon is known as “macro-phase separation,” but IDPs often only undergo this process at protein concentrations greater than what is seen endogenously. IDP heterogenous clustering is a key driver of MLO formation and macro-phase separation, and the molecular mediators of this clustering are often similar or identical to those that occur within MLOs and drive macro-phase separation [3].
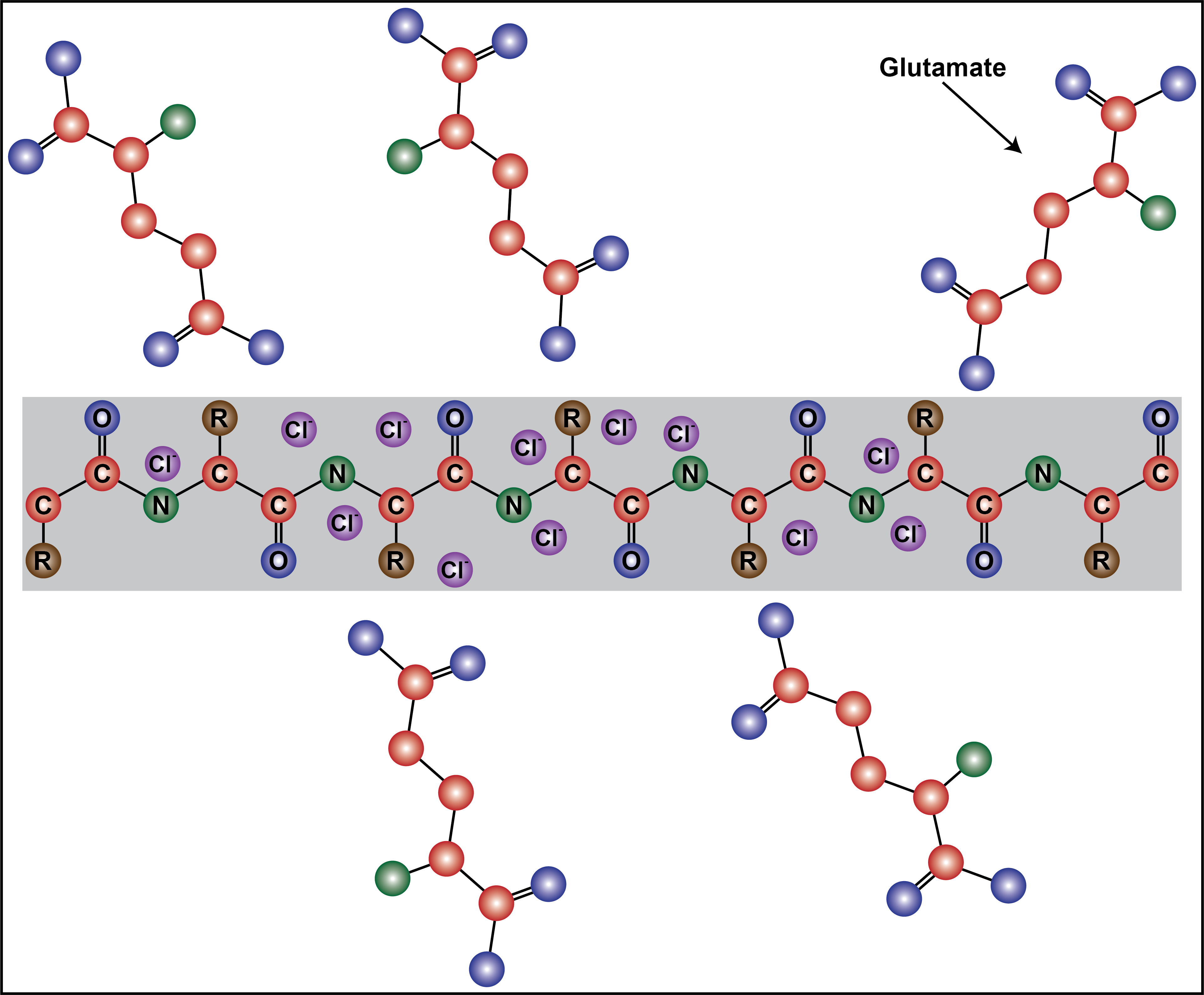
In this preprint, the authors sought to understand the cellular milieu’s contribution to IDP self-association and to distinguish if these environments promote macro-phase separation or clustering interactions. To investigate these questions, a prototypical self-associative IDP, namely FUS, was assayed in solutions containing potassium and glutamate ions (KGlu), which are the most abundant and relevant intracellular ions [4]. The effect of glutamate was of particular interest, because it is a bulky molecule and carries a delocalized negative charge across two carboxylate oxygens, whereas a small ion, such as chloride, contains a strong centralized negative charge. This contributes to glutamate’s relatively weaker shielding effect on cations, causing a less significant reduction on intermolecular interactions, compared to chloride [5]. This work demonstrates the influential role of the cellular milieu on cellular organization and highlights glutamate as a modulator of intermolecular clustering interactions.
Key Findings
Macro-phase separation is minimally affected by glutamate compared to chloride
Using pelleting assays, the concentration at which FUS begins to form phase-separated condensates separate from the bulk solution (saturation concentration or Csat) was determined in KCl and KGlu buffers. These results, coupled with microscopy, showed minor differences in macro-phase separation between KCl and KGlu, with similar Csat values and condensates appearing at nearly the same concentration between the two buffers.
Clustering of FET proteins is increased in KGlu compared to KCl
With macro-phase separation relatively unaffected by the presence of glutamate, the authors sought to investigate the clustering of FUS molecules. For these questions, they employed a number of biophysical approaches. Nanoparticle tracking analysis (NTA) is a microscopy technique that records the Brownian motion of nanoparticle trackers and relates that movement to the size of the particle that the tracker is moving through [6]. In 100 mM KGlu, the authors measured a four-fold increase in the concentration of mesoscale clusters of FUS, compared to 100 mM KCl.
In addition to the NTA experiments, several microscopy techniques were used to confirm the formation of these transient complexes within the sub-saturated solution. These techniques all detected clustering of FUS molecules into complexes at FUS concentrations as low as 400 pM, far lower than endogenous FUS levels [7]. Finally, dynamic light scattering (DLS) was used to confirm the formation of higher-order complexes in proteins from the same family as FUS, namely, EWS and TAF15. These experiments suggest that clustering may be a relevant mode of self-association even when macro-phase separation is not readily occurring.
Tyrosine-arginine interactions between FUS prion-like domain and FUS RNA-binding domain are important mediators of clustering and phase separation
Having established the increased clustering propensity of FUS in KGlu solvents, the authors wanted to understand what interactions mediate this mode of self-association, and if they are similar interactions that drive macro-phase separation. FUS is known to undergo macro-phase separation through interactions between tyrosine residues within the N-terminal prion-like domain (PrLD) and arginine residues in the RNA-binding domain (RBD) [8].
As such, in both KCl and KGlu buffers, tyrosine-to-serine mutations in the PrLD significantly increased the Csat, demonstrating the lower propensity to undergo macro-phase separation. These mutations also made the clustering of FUS molecules undetectable in both solvents, suggesting that these tyrosine-serine interactions also mediate the self-association in sub-saturated solutions. This reduction in macro-phase separation and clustering was also seen when arginine residues in the RBD were mutated to glycine.
When the RBD arginine residues were mutated to lysine, macro-phase separation and clustering were both reduced in KCl buffers. However, interestingly, only macro-phase separation was reduced in KGlu buffers, while clustering was similar to WT. In sub-saturated solutions, clustering is mediated by cation-π interactions, with lysine still able to act as an effective cation to interact with the π system of the tyrosine aromatic rings. However, macro-phase separation is greatly influenced by solubility, which is increased when arginine residues are substituted for lysine. Arginine has a less favorable free energy of hydration compared to lysine, making it behave more hydrophobically and decreasing the solubility of the protein it is incorporated into [9].
Differences between glutamate and chloride in driving protein clustering may be due to the inability of glutamate to interact with backbone amides
Next, the authors turned to molecular dynamics simulations to understand how glutamate’s interaction with FUS differs from that of chloride. These experiments revealed lower interaction coefficients for chloride’s interaction with all amino acid residues, except arginine and lysine, compared to glutamate. These results suggest that the bulkier glutamate anion is able to interact with the side chain of positively charged residues, but is excluded from the peptide backbone, due to its larger size. Chloride, on the other hand, is small enough to associate with the backbone amide, providing a greater charge shielding effect compared to glutamate. This key difference in the interaction between glutamate and chloride with the peptide is likely a vital contributor to the increased clustering seen in KGlu buffers.
Why I chose this study
Understanding the atomic details underlying the interactions driving macromolecular self-association is fundamental to our understanding of how the cell is organized. This preprint drives forward the field of biomolecular and cellular organization by emphasizing the necessity of studying these phenomena in the context of the cellular milieu.
Additionally, the profound effect of solvent properties on the clustering of macromolecules is highlighted by the difference between cellularly abundant glutamate and chloride, which is commonly used for in vitro studies. This preprint identifies key interactions that occur, not just between macromolecules, but between the macromolecules and their cellular environment. This is often overlooked or not given its full due in the context of these biochemical systems.
Questions for the authors
- Based on the increased self-associative clustering seen with glutamate, would you expect to see enhanced or higher affinity interactions between intrinsically disordered proteins, like FUS, and their protein binding partners? This could have implications for the “scaffolding” ability of these highly disordered proteins.
- Other metabolites, which could be in relatively high concentration in particular regions of the cell, could have an outsized effect on the associative properties of molecules in that region. Alternatively, could the formation of these enzymatically-formed metabolites be having a compounding effect on the clustering, coupled to distinct cellular processes?
References
- Wang, B. et al. Liquid-liquid phase separation in human health and diseases. Signal Transduct Target Ther 6, (2021).
- Feric, M. & Misteli, T. Function moves biomolecular condensates in phase space. BioEssays 44, (2022).
- Kar, M. et al. Phase-separating RNA-binding proteins form heterogeneous distributions of clusters in subsaturated solutions. Proc Natl Acad Sci U S A 119, (2022).
- Milo, R., Phillips, R. & Orme, N. Cell Biology by the numbers. (Garland Science, 2015).
- Kozlov, A. G. et al. How Glutamate Promotes Liquid-liquid Phase Separation and DNA Binding Cooperativity of E. coli SSB Protein. J Mol Biol 434, 167562 (2022).
- Filipe, V., Hawe, A. & Jiskoot, W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res 27, 796–810 (2010).
- Patel, A. et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077 (2015).
- Murthy, A. C. et al. Molecular interactions contributing to FUS SYGQ LC-RGG phase separation and co-partitioning with RNA polymerase II heptads. Nat Struct Mol Biol 28, 923–935 (2021).
- Fossat, M. J., Zeng, X. & Pappu, R. V. Uncovering Differences in Hydration Free Energies and Structures for Model Compound Mimics of Charged Side Chains of Amino Acids. Journal of Physical Chemistry B 125, 4148–4161 (2021).
doi: https://doi.org/10.1242/prelights.35670
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Also in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (No Ratings Yet)
(No Ratings Yet)