Integrative Brain Transcriptome Analysis Links Complement Component 4 and HSPA2 to the APOE ε2 Protective Effect in Alzheimer Disease
Posted on: 17 December 2020
Preprint posted on 24 November 2020
'Complement'ing APOE ε2 in Alzheimer’s protection: A meta-analysis of differentially expressed genes in humans.
Selected by Theresa PohlkampCategories: genomics, immunology, neuroscience
Background
Alzheimer’s disease (AD) is a neurodegenerative disease presenting with progressive forgetfulness. AD pathology is defined by deposition of neurotoxic Aβ particles into plaques between brain cells, and neurofibrillary tau-tangles within neurons. The APOE gene encoding the cholesterol transporter Apolipoprotein E is the most significant genetic risk factor for developing the most common, late onset form of AD (LOAD). Of the three common allelic isoforms (ε2, ε3, and ε4), APOE ε3 is most frequent and considered neutral with respect to AD. If you carry one (e.g. ε4/ε3) or two (ε4/ε4) alleles of ε4, your risk to develop AD increases stepwise; contrarily, if you carry one or two alleles of ε2 your risk decreases. Most scientific studies try to uncover the mechanism how APOE4 contributes to AD pathology, less often the focus is on APOE2 and its protective role. In this preprint, Panitch et al. investigate differentially expressed genes (DEGs) between AD cases and controls, with a special emphasis on ε2/ε3-carriers. Their meta-analysis identifies components of the complement signaling pathway as top DEGs, and a glia-enriched APOE ɛ2 related co-expression network including complement pathway genes.
The complement system has been implicated in diverse neurodegenerative conditions, in most scenarios it drives inflammation to exacerbate pathology (Morgan, 2018). The complement cascade comprises several secreted effector components (proteins), their fragments, and membrane-bound complement receptors (CRs). It is part of the innate immune response and contributes to the extent and limit thereof. In general, antibodies recognize antigens, for example presented on pathogens. In the classical complement cascade, the initial antibody-recognition component is within the C1-complex. Activated C1 splits C2 and C4 into their respective a and b parts. C4b and C2a combine to activate C3, whose b-part is recruited to form C4b2a3b, which in turn converts C5. The now converted C5a-part has potent inflammatory and chemotactic activities, and the C5b-part anchors on the target cell surface to recruit and polymerize with C6-C9. Together, they constitute the lytic membrane-attack complex, a cylindrical hole in the antigen-presenting target cell. CRs are mostly expressed on blood and immune cells; upon binding to effector protein fragments, they induce diverse intracellular immune responses. Overall, the complement system can have both beneficial and detrimental effects in various neurodegenerative disorders (Schartz and Tenner, 2020). Notably, CR1, which primarily acts inhibitory upon C3B/C4B-binding, is expressed by brain microglia and CR1 is among the eight highest genetic risk factors for LOAD (Bellenguez et al., 2020; Wightman et al., 2020). In addition, and likely relevant in AD, APOE binds the C1 complex to keep inflammation in check (Yin et al., 2019). Another study found increased expression of several complement-pathway genes in an AD-mouse model encoding human APOE ε3 as compared to ε4 (Fitz et al., 2020). Thus, a link between the AD-protective isoform APOE ε2 and the complement system is an exciting piece to add to the puzzle.
Findings
Panitch and colleagues quantified DEGs in AD and control groups of three gene expression data sets of postmortem brains (ROSMAP with 627, MAYO with 162, and FHS/BUADC with 193 individuals, respectively). When they compared DEGs between AD- and control-groups dependent on APOE genotype (ε2/ε3, ε3/ε3, ε3/ε4), they found transcriptome-wide significant upregulation of C4A, C4B, and GFAP for AD, with the greatest and most significant difference within the ε2/ε3-group (Fig. 1A). When they plotted C4A, C4B, and GFAP expression levels with the progression of tau- (Braak stage) and Aβ- (CERAD score) pathology (ROSMAP dataset), they found that the expression of each of the three genes positively correlated with both pathologies. More specifically, the correlation was significant for all three genes and Braak in the genetic groups ε2/ε3 and ε3/ε3 but not ε3/ε4 (Fig. 1A). Plaque load correlated significantly for all three genes only for ε2/ε3-, but not for ε3/ε3- or ε3/ε4-groups.
In a gene co-expression network enrichment analysis (WGCNA) incorporating all three datasets, they defined 23 networks in total, and one of them (“M01” Fig. 1B) was specific to the APOE ε2/ε3 AD-cohort. Next, they analyzed single nucleus RNA sequencing (snRNAseq) data, which were available from the ROSMAP dataset (FASTQ, 48 subjects), for neuron/glia cell-type specific expression profiles and gene enrichment in the 23 networks. Overall, the individual networks were enriched in specific cell-types, APOE genotype and/or AD/control-groups. However, closer attention was drawn to the M01 network, which contained several complement pathway genes, including C4A, C4B, and CR1, and was enriched in most glia types (astrocytes, but stronger in oligodendrocytes and their progenitor cells) except microglia, but not in neurons. Within M01, 19 genes (including C4A and C4B) correlated with tau- and Aβ-pathologies. Lastly, by analyzing the expression of the most relevant M01 genes with AD-related proteins (within FHS/BUADC samples), they found that C4A, C4B, GFAP, PHLPP1, HSPA2, and DOCK1 positively correlated with phosphorylation of tau but not with Aβ.
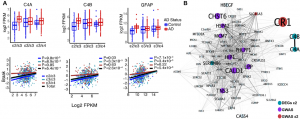
Figure: A) Most significant DEGs between APOE groups (upper graphs) and their expression correlation with tau pathology (lower graphs). B) M01 gene co-expression network in APOE ε2/ε3 AD-group. DEGs in total samples and APOE ε2/ε3 group (turquoise), GWAS AD-risk genes differentially expressed in total samples (purple) and APOE ε2/ε3 group (red). (Reproduced and modified from original preprint, made available by a CC BY-NC-ND 4.0 international license)
Short discussion about the genetic hits GFAP, PHLPP1, HSPA2, and DOCK1:
The glial fibrillary acidic protein (GFAP) is upregulated in reactive astrocytes within the AD brain, which correlates with enrichment and secretion of complement components (Goetzl et al., 2018). PHLPP activation has been described to be dependent on specific CRs during proliferation and RTK-signaling (Strainic et al., 2020). DOCK1 and HSPA2 have not been linked to complement before. While HSPA2 is associated with AD-risk (Lancour et al., 2020), the DOCK1 gene product affects neuronal morphogenesis (Shi, 2013).
What I like about this preprint
I study the function of human APOE isoforms in mice. Recent research highlights that mouse glia in AD-models behave differently from human glia, also with respect to APOE genotype (Tcw et al., 2019). Thus, it is important to keep on track with human APOE isoform function in the species they evolved in and in which they contribute to AD risk. In humans, this preprint identifies an AD-related link between APOE, particularly isoform ε2, and complement pathway genes in glia, most robustly in oligodendrocytes, but not neurons. The enrichment of M01 genes particularly in oligodendrocytes further stresses the question of the involvement of white matter in AD pathology. While AD pathology is most obvious in the gray matter, myelination-deficits in the white matter have been observed (Butt et al., 2019). I appreciate that the authors broadly discuss their study’s weaknesses, for example that the absence of C4A/B-mRNAs in the snRNAseq data is possibly due to their extranuclear localization. They also clarify that their findings do not provide any mechanistical information about the interaction between APOE (ε2) and complement.
Questions for the authors
- In your study you adjusted the data for sex. Did you also look at the data in a sex-dependent manner? If so, are there any interesting insights with respect to complement pathway related genes?
- Several complement pathway related genes were upregulated in ε2/ε3-AD as compared to ε2/ε3-control. Thus, it could be interpreted that APOE ε2 fails to be protective when complement pathway related genes are upregulated. However, in your title and conclusion, you suggest that the complement system may confer a neuroprotective effect against AD through interaction with APOE ε2. Can you elaborate how you got to this conclusion and a scenario how this might work out?
- Given the data would exist, do you think comparing paired data of siblings, in each pair one being ε3/ε3-AD and the other ε3/ε2-control, would help us to identify APOE ε2-related protective gene networks / pathways?
- Have you seen alterations in APOE expression between AD/control, APOE genotype, and/or cell-types?
References
doi: https://doi.org/10.1242/prelights.26492
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the immunology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Scalable transcription factor mapping uncovers the regulatory dynamics of natural and synthetic transcription factors in human T cell states
Inês Caiado
Also in the neuroscience category:
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the immunology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)