Molecular structure of the intact bacterial flagellar basal body
Posted on: 6 April 2021 , updated on: 21 April 2021
Preprint posted on 6 December 2020
Article now published in Nature Microbiology at http://dx.doi.org/10.1038/s41564-021-00895-y
Johnson et al. resolve the high-resolution cryo-EM structure of the intact flagellar basal body from Salmonella
Selected by NYUPeerReviewCategories: biophysics, microbiology
Background
Bacterial and prokaryotic motility often depend on physical appendages called flagella, which protrude from the organism’s membrane and initiate motion through rotational movement, propelling the organism in a directional manner. This movement is important for the organism’s survival and fitness. The flagellar basal body is located at the base of the flagellum, and is important for assembling the flagellum as well as facilitating rotational movement. The basal body is composed of a complex network of transmembrane proteins that form rings and anchor flagellar components to the bacterial inner and outer membranes and the intervening cell wall in Gram-negative bacteria (Figure 1). Flagellar rotation is generated by proton flow from the periplasm through the stator, which rotates the C-ring. Rotation of the C-ring, which is coupled to the axial rod (the portion of the flagellum that transverses the inner and outer membranes and cell wall), drives flagellar motion. Previous genetic and biophysical studies have partially characterized the role of many constituent flagellar components, including rotational dynamics and structural arrangement in the bacterial membrane. Low resolution structures obtained from cryo-electron tomography have identified key complexes that form the basal body, outlining the rough positioning of its components, and high-resolution insights of some basal body subcomplexes such as portions of the axial rod and cap protein are also available. However, a high resolution structure of the entire basal body is not available, and represents a gap in our knowledge of how this large and complex protein machine may function. In this preprint, Johnson et al. resolve the cryo-EM structure of the intact Salmonella flagellar basal body at high resolution (Figure 2). This is a stunning structure of an immensely large multi-protein complex, providing exciting new insights into the structure of the axial rod and the outer membrane “bushing” complex, and how the 173 protein components of the basal body assemble to enable flagellar rotation.
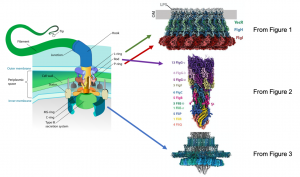
Image credit: Mariana Ruiz Villarreal via Wikipedia (left diagram) and Figures 1, 2, and 3 from Johnson et al.
Key Findings
● Cryo EM structure of the fully assembled flagellar basal body, including the LP-ring, the MS-ring, and the rod proteins at a 2.2-3.7 Å resolution. This includes a ~2.2 Å resolution reconstruction of the OM “bushing” complex, consisting of 26 copies each of FlgH, FlgI, and YecR.
● Identification of a lipoprotein protein YecR, bound to the outer surfaces of the LP-ring, proposed to play a role in the assembly of the LP-ring by modulating interactions with lipids in the surrounding membrane.
● Components of the axial rod and their respective symmetries are resolved. Comparison of this structure of the export gate and associated axial rod with a previously determined cryo-EM structure of an isolated axial rod export gate in the closed state, revealed a possible mechanism by which the gate may open to allow flagellar components to be secreted through the periplasm and outer membrane.
● The intact hook cap complex, which is responsible for organizing the secretion and assembly of hook proteins was resolved. In comparison to previous models suggesting that the cap protein would rotate as new proteins were added to the growing filament, this work suggests that the cap protein is not rotating but rather undergoing a stepping motion tracking the growing tip. The hook cap complex forms a loosely associated pentameric structure that allows for the addition of hook cap proteins to the nascent hook in a “stepped revolution” fashion.
● The detail of this structure improves our understanding of the interactions between the axial rod and ring component interfaces in the basal body. Analysis of these interactions at the molecular level provides an explanation for the discontinuous rotation of the flagellum in 26 discrete steps based upon the 26-fold symmetry of the LP-ring complex.
Why we chose this preprint
While previous studies have characterized individual components of flagellar assembly or have reconstructed large complexes at modest resolution, this structural analysis provides a high resolution map of intact flagellar basal bodies with sufficient detail to hypothesize the mechanism of flagellar assembly and secretion of flagellar components. Understanding modes of secretion and assembly of the flagellum better our understanding of bacterial motility and even pathogenesis. Additionally, achieving such high resolution structures complex molecular machine is impressive and will be of broad interest to the microbiology field.
Questions for the authors
● How does the Salmonella basal body compare to flagella from other bacterial species? Are the core components of this structure broadly conserved?
● Could this work be used for the future development of molecules that target the flagellar basal body assembly and potentially drug development?
● At what point in basal flagellar assembly does a proton gradient drive rotation? At what point are these components sufficiently assembled to enable rotation? For example, can hook and flagellin assembly occur while the system is rotating?
● What was the most challenging aspect of this study?
References
- Andreas Diepold and Judith P. Armitage. Type III secretion systems: the bacterial flagellum and the injectisome. Philos Trans R Soc Lond B Biol Sci. 370:20150020 (2015)
- Takashi Fujii, Takayuki Kato, Koichi D. Hiraoka, Tomoko Miyata, Tohru Minamino, Fabienne F. V. Chevance, Kelly T. Hughes & Keiichi Namba. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Comm 8:14276 (2017)
- Yumiko Saijo-Hamano, Hideyuki Matsunami, Keiichi Namba, Katsumi Imada. Architecture of the Bacterial Flagellar Distal Rod and Hook of Salmonella. Biomolecules. 9:260. (2019)
- Mariana Ruiz Villarreal, Flagellum diagram, https://en.wikipedia.org/wiki/Flagellum
- Steven Johnson, Emily J. Furlong, Justin C. Deme, Ashley L. Nord, Joseph Caesar, Fabienne F.V. Chevance, Richard M. Berry, Kelly T. Hughes, Susan M. Lea. Molecular structure of the intact bacterial flagellar basal body.bioRxiv 2020.12.05.413195; doi: https://doi.org/10.1101/2020.12.05.413195
doi: Pending
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
Also in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)