Prospective, brain-wide labeling of neuronal subclasses with enhancer-driven AAVs
Posted on: 27 February 2019
Preprint posted on 31 January 2019
Towards precise targeting of single cell types: enhancer-driven gene expression and engineered viral vectors to improve specificity
Selected by Jesus VictorinoCategories: genomics, neuroscience
Background & Summary:
Cellular subclasses have long been studied in order to identify specific features that can be targeted for a wide range of research purposes. Gene expression profiles are a hallmark of cell identity and subpopulations of cells inside a tissue show unique signatures at the transcriptional and protein levels due, in part, to the differential activity of regulatory elements that control gene expression, such as promoters or enhancers.
Driver lines are often used in mouse genetics to label cell populations, although many of these drivers label different cell types in the organism. In this work, Lucas T. Graybuck et al. identified the enhancer repertoire of neuron subclasses based on single-cell (sc) RNA-seq and scATAC-seq. Adeno-Associated Virus (AAV) vectors capable of infecting the Central Nervous System (CNS) combined with specific enhancer elements effectively drove the expression of the transgene in one out of the three neuron subclasses located at the layer 5 (L5) of the visual cortex of the brain, improving specificity of existing mouse driver lines.
Key findings:
The authors first characterized open chromatin regions by scATACseq of >2,400 cells from the brain by using 25 driver lines. Combination with scRNAseq showed an increase in cell type resolution of brain subpopulations and data integration identified putative enhancers of Fam84b and Hsd11b1 genes specifically accesible in L5 excitatory (L5-PT) and intertelencephalic (L5-IT) neurons, respectively.
Once the authors identified candidate regulatory elements for the neuron subclasses, they cloned their sequences into Adeno-Associated Virus (AAV) vectors and combined enhancer specificity with AAV tropism to effectively target the cells of interest. They took advantage of the AAV-PHP-eB engineered serotype that is able to cross the blood-brain barrier and infect the central nervous system (CNS) after intravenous injection [1, 2]. Viruses containing a fluorescent reporter controlled by a minimal promoter together with the enhancer of interest were tested. In the case of the putative regulatory element 4 (mscRE4) controlling the Fam84b gene, they achieved >91% targeting of pyramidal L5 excitatory neurons (L5-PT), without labeling intertelencephalic neurons (IT) from the same layer of visual cortex (Fig. 1).
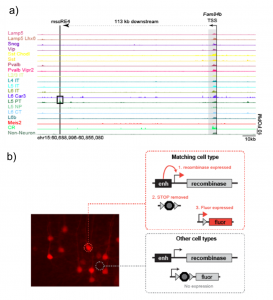
Figure 1. mscRE4 enhancer in the Fam84b locus is specific of L5-PT neurons (a) (image taken from figure 2e) and its use to control recombinase gene expression in combination with AAV vectors (b) (taken from figure 1) resulted in resolved cell subclasses.
To expand the use of AAV-mediated enhancer-driven gene expression and solve the limitations of the available mouse lines, they applied this strategy to express recombinases in the neurons of interest. Co-administration of viruses carrying different enhancer-recombinase pairsincreased the versatility of the tool and enabled simultaneous labelling of cell populations without the need for multiple crosses between lines.
Why I chose this work:
Distinguishing and characterizing cell types is not only essential to elucidate the role they are playing in a fully developed organ, but also to understand how an organism develops from a single cell. Specific labelling of cell populations allows for their isolation to study particular features such as their transcriptomic profile or other genomic properties like chromatin accessibility, histone marks or transcription factor binding. Fluorescently labelled populations can also be tracked down during embryo development to better understand the contribution and the dynamics of the different progenitors and visualize the formation of complex structures such as the nervous system or the heart. For this, again, we depend on already available recombinase-based driver lines that might not label our tissue of interest or have a spread effect that would turn the analysis rather difficult.
I chose this preprint for its elegant experimental design based on single-cell characterization of brain subpopulations which enabled the identification of specific enhancers that can be used to target and/or label cells with higher precision. This work will be of interest not only to neurobiologists who will find a useful resource of brain genomics, but for many developmental biologists who will also find a powerful strategy to increase the resolution of lineage tracing.
AAVs are being widely assayed in clinical trials for their presumably undetectable immune response and delivery potential. Surface markers, such as receptors or epitopes, provide a collection of gates through which exogenous DNA can be introduced into the desired cell by means of different delivery systems, as in the case of viral vectors. To meet sensitivity and specificity, which are crucial for successful practice, identifying surface markers that are present in the population of interest is as important as their absence in others that should be kept non-transduced. However, it is not always possible to meet these requirements and target exclusively the tissue under study due to shared features between cell types and limitations in the delivery system. The strategy followed by Lucas T. Graybuck and collaborators, combining AAV tropism and enhancer specificity could also be of great interest in the field of gene therapy to reduce off-target effects.
Questions to the authors:
- When we take a look at the cell labelling by mscRE4 and mscRE16 in figure 4, we can see fluorescent cells beyond those of layer 5 indicative of spread labelling throughout the brain:
- Are there other labelled regions in the rest of the organism?
- Do the authors think the system could be improved in terms of specificity (those labelled populations in the brain outside the L5) by using a 2 components system each one controlled by a different enhancer?
- Since accessible chromatin is assayed at the single cell level and there are not multiple copies of the same region per cell (contrary to what happens with transcripts):
- How reproducible open chromatin profiles are in the scATAC-seq data between cells within the same transcriptomic cluster?
- Are scATAC-seq profiles composed of a portion of the profile of population ATACseq? And if so, once the profiles of several replicates are overlapped one could get a picture of the population profile. What is the estimate percentage of open regions that is captured by a single cell?
References:
- Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, Wu WL, Yang B, Huber N, Pasca SP, Gradinaru V. 2016. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nature Biotech. 34, 204–209.
- Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sánchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, Gradinaru V. 2017. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci. 20, 1172-1179.
doi: https://doi.org/10.1242/prelights.9000
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the neuroscience category:
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)