Single molecule mechanics resolves the earliest events in force generation by cardiac myosin
Posted on: 21 August 2019
Preprint posted on 27 June 2019
Article now published in eLife at http://dx.doi.org/10.7554/eLife.49266
Caught in the act: measuring cardiac myosin's catalytic cycle at high spatial and temporal resolution
Selected by Alyson SmithCategories: biochemistry, biophysics
Why I think this study is interesting
Myosin uses a cycle of ATP hydrolysis coupled to conformational changes to pull on actin filaments and sustain eukaryotic life. This study uses ultra-fast force clamping, a sensitive biophysical assay, to capture the fastest events in myosin’s ATPase cycle, which have remained opaque to previous assays. The study’s results provide insight into the inner workings of the myosin motor and can pinpoint problems underlying human diseases caused by myosin mutations, such as cardiomyopathy, muscular dystrophy, or cancer.
Background
Most of the dozens of myosin isoforms—including skeletal, cardiac, smooth muscle, and nonmuscle—follow the force-generating cycle diagrammed below (1). First (steps A and B), myosin motor domain heads hydrolyze ATP to ADP and inorganic phosphate (Pi). At this stage, myosin heads bind weakly to actin filaments. Next (step C), myosin releases phosphate. At around the same time, myosin’s lever arm swings to produce the force-generating power stroke (termed working stroke in this preprint). Finally (step D), myosin heads exchange ADP for ATP and release actin to restart the cycle.
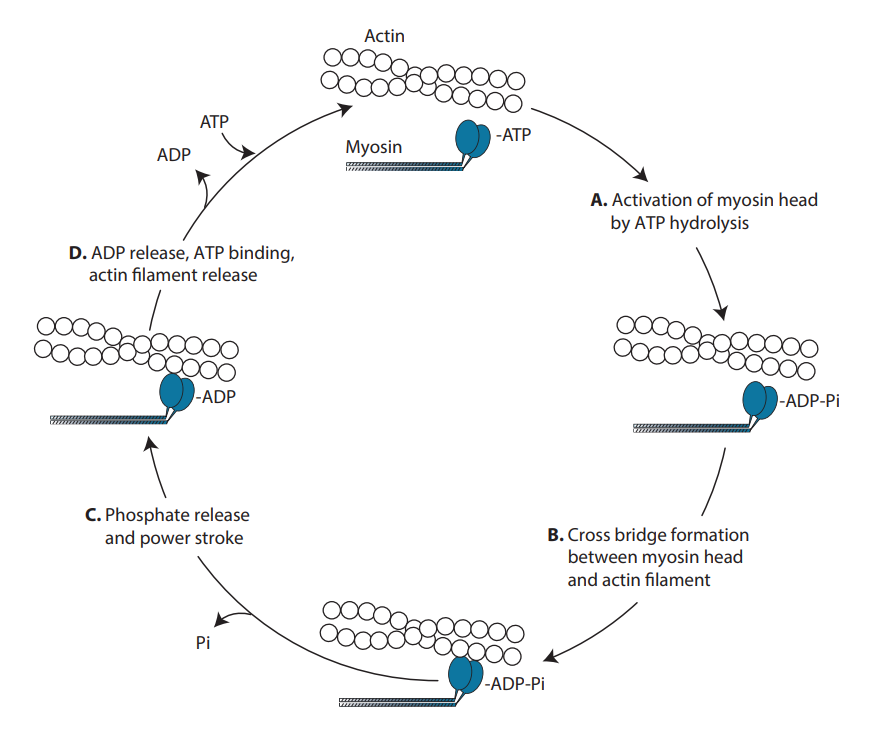
Following actin binding by myosin motor domains (step B), conformational changes in myosin and actin, the swinging of the lever arm, and phosphate release occur within milliseconds, making each event difficult to resolve with conventional biophysical methods. This study zooms in on these events to determine which occurs first for human beta-cardiac myosin: the power stroke or phosphate release.
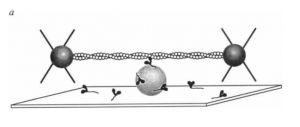
To achieve high temporal resolution, the authors used an ultra-fast force clamp with a three-bead optical trap (2). Two laser-controlled beads moved an actin filament across a coverslip holding myosin molecules attached to beads. When the actin encountered a myosin head, force feedback loops applied to the beads detected sub-nanometer displacements of the actin at intervals of less than 100 microseconds.
The high resolution of ultra-fast force clamping produced traces of myosin-actin interactions filled with fluctuations due to Brownian motion. This variability precluded applying computational techniques (step finding algorithms, hidden Markov chains, or Bayesian non-parametric analysis) to individual traces. The authors instead used ensemble averages of many traces, aligned at the beginning of the myosin-actin interactions, to detect the power stroke and other events in their experiments.
Key findings
Many myosin-actin interactions are short-lived
Many myosin-actin interactions ended before myosin completed its ATPase cycle. The frequency of these short-lived interactions increased with applied load; up to 50% lasted less than 10 milliseconds at the highest load (4.5 picoNewtons). Despite their transience, short-lived interactions stopped the laser-powered motion of the actin and held it for hundreds of microseconds. The direction of the applied load affected the relationship between force and interaction duration. This polarity indicates that the short-lived state represents a weak but stereospecific interaction between actin and myosin that either ends quickly or proceeds to a strong-binding state and force generation.
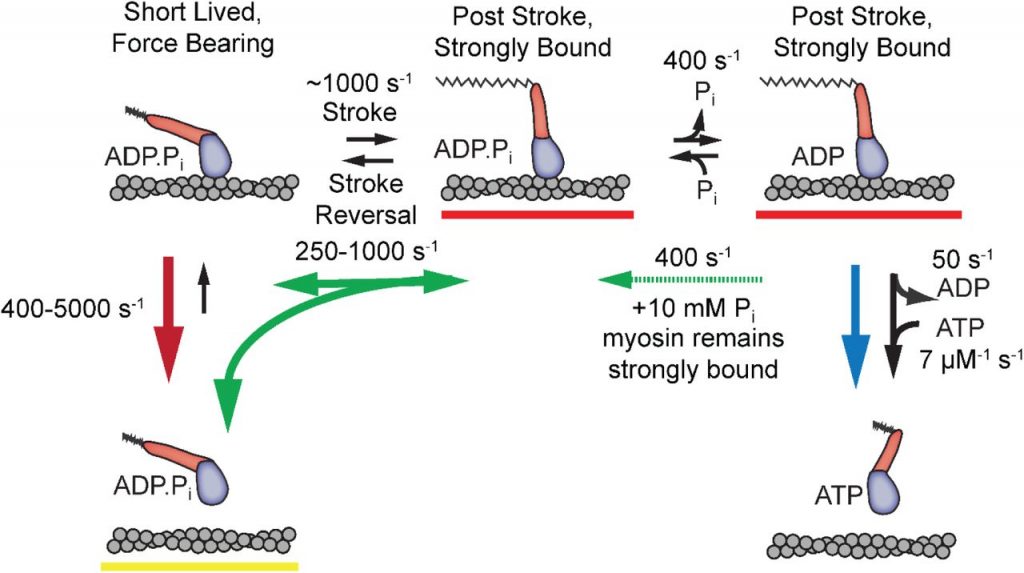
The power stroke occurs before phosphate release
The ultra-fast force clamp detected myosin’s power stroke as a small displacement that occurred within 5 milliseconds of actin binding. The stroke happened faster than both myosin-actin detachment and phosphate release, making it unlikely that phosphate release occurs before the power stroke. Ten millimolar free phosphate did not change the stroke rate, further supporting the author’s model that the power stroke occurs before phosphate release.
The power stroke and phosphate release are reversible
After the power stroke reached its peak actin displacement, the authors saw small displacements in the opposite direction. Adding free phosphate to the assay amplified this reversal and decreased the final displacement. This displacement reversal indicates that free phosphate can re-bind after release, slowing myosin’s ATPase cycle. The fact that free phosphate affects post-stroke displacement reversal but not the power stroke itself further supports the author’s hypothesized order of events: power stroke first and phosphate release second.
Future directions
According to the author’s data and simulations, myosin remains bound to actin in a pre-stroke state for ~300 microseconds before a stroke reversal but ~1 millisecond following a reversal. This increased state duration suggests that myosin does not return to its original conformation following phosphate re-binding and stroke reversal. Future simulations and biophysical experiments could test this hypothesis.
The results of this study agree with previous work on skeletal and cardiac myosin, non-muscle myosin V, and ultra-fast force clamp experiments using fast-skeletal muscle myosin (see references in preprint). The dozens of eukaryotic myosin isoforms differ in sequence, structure, kinetics, and biomechanics (3). Further research using ultra-fast force clamps or other sensitive biophysical techniques may reveal different mechanisms for myosin force generation.
How do mutations in the myosin motor domain affect the power stroke and phosphate release? Disease-causing mutations can occur in the actin-myosin interface, in the ATP/ADP binding site of myosin, or in myosin domains required to support conformational changes. Ultra-fast force clamp assays could determine how these mutations affect the power stroke and phosphate release and whether potential therapeutics lessen their effects.
References
1. Kaplinsky E and Mallarkey G. Cardiac myosin activators for heart failure therapy: focus on omecamtiv mecarbil. Drugs Context 7:212518 (2018).
2. Finer JT, Simmons, RM, and Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature 368, 113–119 (1994).
3. Sweeney HL and Houdusse A. Structural and functional insights into the Myosin motor mechanism. Annu Rev Biophys 39:539-557 (2010).
doi: https://doi.org/10.1242/prelights.13537
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Also in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (1 votes)
(1 votes)