Building customizable auto-luminescent luciferase-based reporters in plants
Posted on: 15 November 2019
Preprint posted on 17 October 2019
Article now published in eLife at http://dx.doi.org/10.7554/eLife.52786
Night-light plants! A fungal bioluminescence pathway adapted for use in plants results in an auto-luminescent reporter system.
Selected by Martin Balcerowicz, Jonny CoatesCategories: bioengineering, plant biology, synthetic biology
Context and background
Bioluminescent reporters such as the firefly luciferase have been widely used in eukaryotic systems. They are particularly appealing for use in plants as they do not require excitation and hence autofluorescence that is inherent to plant tissue can be avoided. A major disadvantage however is the need to deliver the luciferase substrate exogenously – causing issues with homogenous substrate delivery and tissue penetration. It would thus be highly desirable to generate plants that already produce the luciferase substrate. Kotlobay et al. (1) identified a novel luciferase/luciferin biosynthesis system, referred to as the caffeic acid cycle, in the eukaryotic fungus Neonothopanus nambi (Fig. 1). The fungal luciferin 3-hydroxyhispidin is produced from caffeic acid through the activity of two biosynthetic enzymes, hispidin synthase (HispS) and hispidin-3-hydroxylase (H3H). The luciferase (luz) subsequently generates a bioluminescent signal by oxidising 3-hydroxyhispidin; the resulting product can be recycled to caffeic acid by caffeoylpyruvate hydrolase (CPH). In their preprints, Mitiouchkina et al. and Khakhar et al. deploy this system in plants and show that it can be harnessed to monitor gene expression.
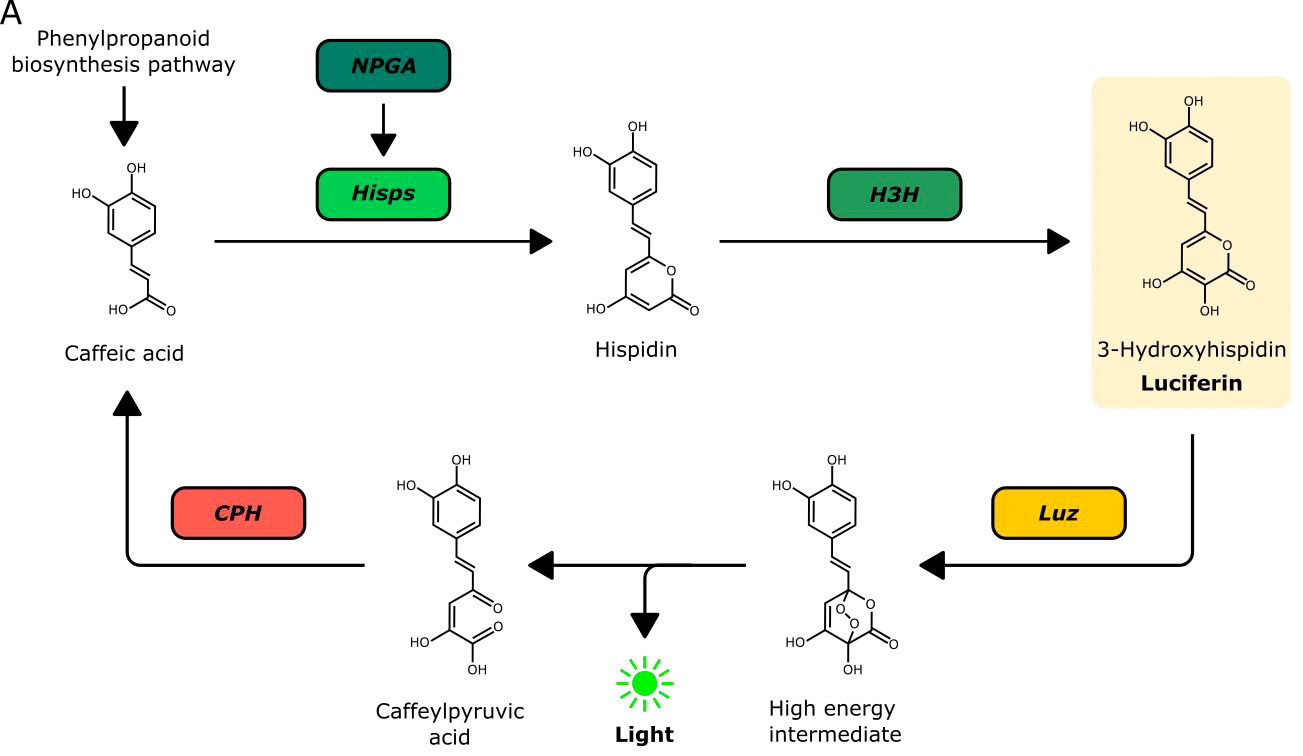
Figure 1. Fungal bioluminescent pathway (caffeic acid cycle) used to create bioluminescent plants. Reproduced from Figure 1 of the preprint (Khakhar et al.) under a CC BY-NC-ND 4.0 license.
Key findings
- The fungal caffeic acid cycle effectively labels multiple plant species without the need for lasers or other light sources
Caffeic acid is an intermediate of the plant’s phenylpropanoid biosynthesis pathway and present in virtually all plant species. The authors of both preprints introduced genes encoding the enzymes of the fungal caffeic acid cycle into plants to test whether the pathway can be reconstituted in planta, thereby bypassing the need to externally supply luciferin. Transient expression of the enzymes in multiple plant species including tobacco, Arabidopsis and tomato resulted in auto-luminescence of the transformed organs. When the pathway was stably integrated into the tobacco genome, auto-luminescence was observed across the whole plant and throughout development. Mitiouchkina et al. further observed that luminescence varied across organs – over the time of day and strongly increased upon wounding – likely correlating with the activity of the phenylpropanoid pathway that generates caffeic acid.
- This system can be used to visualise spatio-temporal changes in gene expression
The luminescence pathway developed in these preprints potentially represents a powerful reporter system. Khakhar et al. adapted this system to observe spatio-temporal changes in gene expression by expressing the fungal luciferase from selected promoters. Expression from the Petunia ODORANT1 promoter lead to exclusive auto-luminescence in flowers with a diurnal rhythm that peaked at dusk, in agreement with previous observations. Expression from the abscisic acid (ABA)-responsive AtRAB18 promoter on the other hand conferred strong increase in luminescence when plants were treated with ABA or dessicated, a stress known to boost ABA levels. None of these specific responses were observed when the luciferase was expressed from the constitutive 35S promoter, providing proof of concept that the fungal bioluminescence pathway (FBP) can be employed as a reporter system for gene expression.

Why we chose this paper
Many model organisms or cell lines are highly amenable to bioluminescence reporter systems such as the firefly luciferase. However, a major disadvantage of these systems is the requirement of exogenous substrate, particularly in plants where tissue permeability varies greatly. The preprints presented here represent an excellent solution to this problem by negating the need for exogenous substrate whilst providing a powerful and adaptable reporter system.
Open questions
- The authors of both preprints highlight the excellent use of this system as a tool within plant biology. How applicable is this reporter system in different plant groups (monocots, ferns, bryophytes)? Is this also applicable to other organisms and do the authors envisage adoption in different systems?
- To what extent may the observed variations in the phenylpropanoid pathway activity affect FBP-based reporter systems? Are there tissues/conditions that might be more suitable for analyses with FBP than others?
- How stable is the expression system across plant generations? Has silencing of these genes in subsequent generations been observed?
References
- Kotlobay AA, Sarkisyan KS, Mokrushina YA, Marcet-Houben M, Serebrovskaya EO, Markina NM, et al. Genetically encodable bioluminescent system from fungi. Proc Natl Acad Sci. 2018 Dec 11;115(50):12728–32.
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the plant biology category:
Actin Counters Geometry to Guide Plant Cell Division
Jeny Jose
The nucleus follows an internal cellular scale during polarized root hair cell development
Jeny Jose
Conservation and divergence of regulatory architecture in nitrate-responsive plant gene circuits
Jeny Jose
Also in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the plant biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)