Tsetse salivary glycoproteins are modified with paucimannosidic N-glycans, are recognised by C-type lectins and bind to trypanosomes
Posted on: 13 July 2020
Preprint posted on 27 June 2020
Article now published in PLOS Neglected Tropical Diseases at http://dx.doi.org/10.1371/journal.pntd.0009071
Categories: biochemistry
Background
Hematophagous insects have evolved special adaptations to ensure a successful bloodmeal from vertebrate hosts, key among which is their saliva. Salivary components have been studied in a range of contexts including interactions with the host immune system. However, few studies have addressed the importance of the post-translational modifications in these proteins. N-glycosylation is a highly common post- and co-translational modification that can affect protein folding, protein stability, ligand binding, and protein antigenicity. N-glycans have a wide variety of functions, encompassing structural and modulatory properties to the binding of other proteins and cell-cell interactions.
African sleeping sickness is caused by Trypanosoma brucei, a parasite transmitted by the bite of a tsetse fly. Trypanosome infection induces a severe transcriptional downregulation of the tsetse genes that encode for salivary proteins, affecting its anti-hemostatic and anti-clotting properties (1). To better understand trypanosome transmission and the potential role of gylcans in insect blood-feeding, in the present work, Kozak et al (2) characterize the salivary glycome of T. brucei-infected and uninfected tsetse flies (Glossina spp.).
Key findings and developments
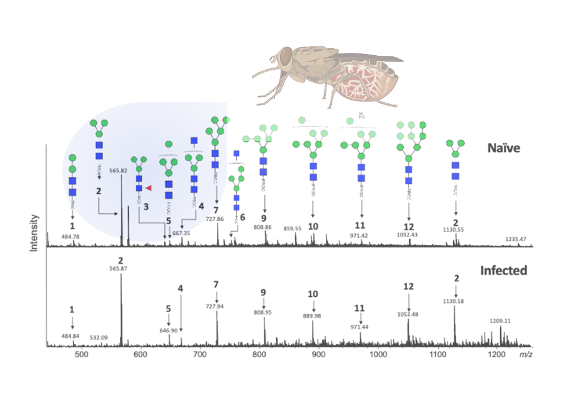
Bioinformatic analysis identified that 72% of Glossina proteins have at least one potential glycosylation site. Further confirmation was performed by molecular methods, and salivary proteins were identified using mass spectrometry. This was followed by identification of the type of glycosylation (N– or O-linked), and proteomic analysis, which showed that suggesting that the main type of sugars linked to tsetse salivary glycoproteins are N-glycans. Characterization of the type of N-glycosylation present indicated the presence of several high mannose or hybrid type N-glycans.
A full structural characterization of G. morsitans salivary N-glycans was then performed. Hydrophilic interaction lipid chromatography (HILIC)- ultra-high pressure liquid chromatography (UHPLC) analysis revealed 13 peaks that correspond to a variety of potential high mannose and hybrid N-glycan structures. The peak of highest intensity corresponds to the core structure Man3GlcNAc2-2AB (paucimmanose). Secondary confirmation of the salivary N-glycan structures was performed by positive-ion ESI-MS and ESI-MS/MS. Overall, these results confirm the findings by HILIC-UHPLC, and suggest that tsetse salivary glycans consist mainly of highly processed Man3GlcNAc2 in addition to several other paucimannose, oligomannose, and three hybrid-type glycans: Man3GlcNAc3, Man4GlcNAc3 and Man5GlcNAc3 .
Since T. brucei infection affects the composition of tsetse saliva (1), the authors investigated if infection changes salivary glycosylation as well. For this, they first compared the salivary profiles of uninfected flies with those that had either a salivary gland or a midgut infection with T. b. brucei. They found no major changes in the profile of salivary proteins in the different physiological states. They then went on to determine whether T. b. brucei infection or fly age (teneral vs. adult) alters the structure of salivary N-glycans, and found no significant quantitative differences between all conditions. They then determined whether trypanosome infection alters the immune reactivity of tsetse salivary glycoproteins. Recognition of control G. morsitans saliva before and after cleavage of the glycans appears unaffected; however, during salivary gland infection the polyclonal serum only detected the high molecular weight proteins after glycans were cleaved. The effect is more readily seen during salivary gland infections, possibly due to the downregulation of other salivary proteins during infection, and seems to be concealed both in the saliva of naïve flies and those with midgut infection. The authors were also able to identify proteolytic products of salivary proteins that are formed as a result of the trypanosome infection in the gland.
Next, they investigated whether glycosylated salivary proteins bind onto metacyclic trypanosomes. Molecular and mass-spectrometry based assays identified components that likely correspond to 5’Nucleotidase-related protein sgp3, TSGF and the Tsal glycoproteins, plus two small additional bands identified using anti-saliva IgGs. Interestingly, the abundant, non-glycosylated TAg5 protein in saliva was found not to bind to the metacylic trypanosome surface. Knowing that N-glycans from G. morsitans salivary glycoproteins are recognised by the mannose receptor and DC-SIGN, the authors performed overlay assays of recombinant mannose receptor and DC-SIGN with tsetse saliva. They found that the mannose receptor recognizes saliva glycoproteins better than DC-SIGN; that recognition is fully linked to N-linked mannosylated glycans, as binding disappears upon treatment with N-glycanase.
Altogether, the authors suggest that although the repertoire of tsetse salivary N-glycans does not change during a trypanosome infection, the interactions with mannosylated glycoproteins may influence parasite transmission into the vertebrate host.
What I like about this preprint
I think the work is very relevant to the field of host-pathogen interactions, and covers a relatively unknown and understudied field in vector biology, which is nonetheless important for our understanding of pathogen transmission. And also on its own, to know more about the differences in saliva among insects!
Questions to authors
- You mention along your work that trypanosomes causes a profound transcriptional downregulation of most tsetse salivary proteins. At what point of infection does this occur? Is this specific to Trypanosoma brucei? If not, can you expand on why this is the case?
- Is it known if fly gender affects their brucei transmission ability, and saliva composition?
- Is it known if salivary content recognition is equal among mammalian species, suggesting perhaps a more successful adaptation of protein contents to one host species but not others?
- You found that only glycosylated salivary proteins associate with the trypanosome surface. Could you expand further on the significance of this? Has something similar been observed in other hosts/pathogens?
References
- Van Den Abbeele J et al: The Glossina morsitans tsetse fly saliva: general characteristics and identification of novel salivary proteins. Insect Biochem Mol Biol, 2007.
- Kozak RP, et al, Tsetse salivary glycoproteins are modified with paucimannosidic N-glycans, are recognized by C-type lectins and bind to trypanosomes, bioRxiv,2020.
doi: https://doi.org/10.1242/prelights.22976
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)