Deep learning-enhanced light-field imaging with continuous validation
Posted on: 4 November 2020
Preprint posted on 31 July 2020
Article now published in Nature Methods at http://dx.doi.org/10.1038/s41592-021-01136-0
Improving the toolkit for 3D imaging with deep learning: HyLFM and HyLFM-Net
Selected by Mariana De NizCategories: bioengineering, physiology
Background
Capturing highly dynamic physiological processes happening on milli-second time scales across large areas in living organisms requires imaging methods capable of such resolution. An attractive candidate for high-speed 3D image acquisition is light-field microscopy (LFM), which has already opened new avenues in the fields of neurobiology and cardiovascular dynamics. While some technical hindrances of this tool have been overcome since its conception, the widespread use of LFM has been hampered by a computationally demanding, iterative image reconstruction process that requires a complex computational infrastructure and adequate data management. Multiple algorithms derived from deep learning and convolutional neural networks have recently been proposed to replace iterative deconvolution procedures, and offer new methods for deblurring, denoising and super-resolution. While many of these methods have excellent performance in various biologically relevant settings, many are not optimal for dynamic imaging with LFM given the complexity of dynamic processes in small animals. In their work, Wagner and Beuttenmueller et al (1) present a novel framework consisting of a hybrid light-field light-sheet microscope (HyLFM) and deep-learning-based volume reconstruction. In it, single light-sheet acquisitions continuously serve as training data and validation for the convolutional neural network (termed HyLFM-Net) reconstructing the LFM volume.
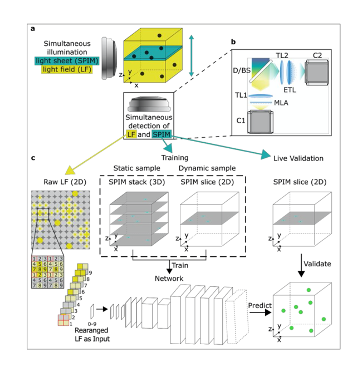
Key findings and developments
A simultaneous selective-plane illumination microscopy (SPIM) modality was added into a standard LFM microscope, allowing the generation of high-resolution ground truth images of single planes for validation, training, and refinement for the convolutional neural network. Training can be done on static sample volumes, or dynamically from a single SPIM plane going through the volume during 3D image acquisition. An automated image processing pipeline ensures that LFM and SPIM volumes are co-registered in a common reference volume and coordinate system with high precision. This is important for convolutional neural network training and validation, and the systems’ ability to acquire 2D and 3D training data is key for reliable convolutional neural network reconstructions, including data never seen in training. Altogether, the HyLFM-Net is trained on pairs of SPIM-LFM images.
To evaluate the performance of the HyLFM system, the authors imaged sub-diffraction sized fluorescent beads suspended in agarose, and quantified the improvement in spatial resolution and image quality compared to standard iterative light-field deconvolution. They concluded that HyLFM-Net correctly inferred the 3D imaging volume from the raw light-field data, with better resolution than that which could be obtained by light field deconvolution, and without artifacts commonly found in light field deconvolution. The authors point out the importance of training on diverse datasets, to avoid biases in performance.
As proof of principle, the authors explore the capabilities of the HyLFM system by imaging the dynamics of a hatchling medaka fish heart in vivo, to demonstrate the capability of the system to correctly capture dynamic cellular movements in 3D. The HyLFM-Net allowed acquiring high image quality metrics compared to SPIM, and allowed 3D volume inference at up to 18Hz, with at least 1000-fold reconstruction speed compared to light field deconvolution. The authors note that the network trained on dynamically acquired SPIM single planes performed equally well or better than the network trained on fully static volumes.
The authors also tested the HyLFM system on transgenic larval zebrafish brains expressing calcium indicators, to monitor neural activity. The ground truth data enabled the HyLFM system to faithfully learn and infer structural, as well as intensity-based information. The authors conclude HyLFM is thus an attractive method for visualizing neural activity.
Altogether, the new system allows reconstructing light-field volumes at sub-second rates, eliminating the main computational hindrances for light field imaging. Moreover, the system enables acquiring appropriate training data simultaneously and on-the-fly, while allowing continuous validation and fine-tuning. The network over time learns on the actual experimental data, rather than requiring pre-acquisition of training images in separate microscopes, solving the hindrance of transferability.
What I like about this preprint
I like that the authors address a hugely important technical gap in a fast-advancing microscopy area. It has not been uncommon over the past few decades that major advances in microscopy tools occur, and the methods for image analysis stay behind. This at some point becomes a limiting factor in itself. I think that tools addressing this gap to overcome those limitations are key, and ultimately allow the different microscopy tools to be used to their full potential, and become widespread.
References
1. Wagner N, Beuttenmueller F, et al, Deep learning-enganced light-field imaging with continuous validation, bioRxiv, 2020.
doi: https://doi.org/10.1242/prelights.25619
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the physiology category:
Trade-offs between surviving and thriving: A careful balance of physiological limitations and reproductive effort under thermal stress
Tshepiso Majelantle
Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Muhammed Sinan Malik
Wide-ranging behavioral dysfunction in two mouse models of pathological human variants in the GRIK2 kainate receptor gene
Pushpinder Singh
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the physiology category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |











 (No Ratings Yet)
(No Ratings Yet)