A conserved MFS orchestrates a subset of O-glycosylation to facilitate macrophage dissemination and tissue invasion
Posted on: 23 January 2019 , updated on: 27 January 2019
Preprint posted on 12 September 2018
Article now published in eLife at http://dx.doi.org/10.7554/eLife.41801
Minerva instructs Drosophila embryonic macrophages to push the boundaries of tissue invasion by initiating a developmentally-controlled program of glycosylation.
Selected by Giuliana ClementeCategories: cancer biology
Context and Background:
Post-translational modifications (PTMs) are covalent modifications of proteins that occur after synthesis and folding and are used by cells to diversify and fine-tune protein activity. As such, PTMs regulate a broad range of events including protein subcellular localization, protein-protein interaction, enzymatic activity, signal transduction and protein degradation. Therefore, it is not surprising that uncontrolled changes in the levels and/or repertoire of PTMs can result in a deregulation of cellular functions which ultimately contributes to aging and diseases.
Glycosylation (i.e. the addition of sugar to the oxygen (O) or nitrogen (N) of amino acid residues) is one of the most abundant post-translational modifications. O-glycosylation is a multi-step process which can result in the generation of truncated O-glycans known as Tn and T-antigens. While at the embryonic stage T and Tn modifications are transient and mostly lost in adults, their de-novo synthesis is often switched back on in cancer cells which therefore display high levels of T and Tn-antigens on their surface. T and Tn-antigens are indeed counted as tumour biomarkers and their presence often correlates with poor prognosis and the appearance of metastatic lesions.
As T and Tn antigens are mostly absent in adult tissues and transiently appear during embryonic development, it is reasonable to draw the conclusion that the appearance of these structures in cancer cells might arise from the reactivation of an embryonic molecular program. Understanding the basis of how O-glycosylation is modulated and controlled during development could therefore crucial to gain a better understanding of cancer biology and to potentially identify new therapeutic targets.
Drosophila macrophages (also known as hemocytes) originate in the head mesoderm and from their site of origin they actively migrate to disseminate throughout the embryo (Fig. 1). During their dissemination, these cells mostly undertake non-invasive routes, such as along the developing ventral nerve cord. However, at the earlier stages of their developmentally controlled migration, these cells breach the epithelial barrier and invade the juxtaposed ectoderm (germband) before its anterior-to-posterior retraction starts (Fig.1 A). For this characteristic migratory behavior, that closely resembles the one of metastatic cells, Drosophila hemocytes offer the unique possibility to verify whether O-glycosylation contributes to tissue invasion and cell dissemination in vivo during embryonic development.
Key points:
A screening of FITC-labelled lectins showed that glycan structures are present on the surface of invading hemocytes. As proof of principle, downregulation of the core T-synthase C1GalTA (which results in a reduction of T-antigen levels) causes an increase in macrophages which sit in the yolk and are unable to invade the germband. Therefore, T O-glycans indeed facilitate germband infiltration (Fig.1 C). What players contribute to an increase in T O-glycans? Given that neither the T-synthase or the sugar transporter Ugalt are transcriptionally upregulated at stage 12, the authors ran an in silico analysis for potential sugar binding protein expressed in macrophages at the time of the increased O-glycosylation and identified CG8602 as a gene codifying for a Major Facilitator superfamily (MFS) protein with predicted functions of sugar transporter. The authors named this protein Minerva.
minerva is upregulated in macrophages at stage 10-12 of development (12-fold increase in hemocytes at stage 12 compared to the rest of the embryo) and its mutation caused a reduction in the T-antigen levels on macrophages at the same developmental stage. Minerva co-localises with the Golgi markers Golgin 84 and with the endosome markers Rab7, Rab11 and Hrs. Most importantly, minerva mutants showed germband invasion defects comparable to the ones cause by C1GalTA RNAi. Analysis of macrophage migratory ability revealed that minerva mutant macrophages moved at slower speed from their initial position to the germband,paused longer to gain entry into the tissue and also, once they infiltrate the ectoderm, they moved slower within the germband. Importantly, hemocyte migration along the ventral nerve cord is not perturbed, arguing against the possibility that the phenotypes observed were due to general defects in developmental dispersal.
To identify what proteins are targeted for glycosylation in a Minerva-dependent manner, lectin-enriched O-glycoproteomics was performed in stage 11-12 wild type versus minerva mutant. Analysis of wild type embryos led to the identification of 270 glycoproteins and revealed that O-glycosylation on Threonine is the most abundant form. O-glycosylated proteins fall in the GO term categories of metabolism, cuticle development and receptors. The O-glycoproteome of minerva mutants mostly overlapped with the wild type one with only a small subset of proteins (63 proteins, 23%) showing a 3-fold change and 18 proteins (6%) a ten-fold change. Of these 18 proteins with lower levels of T antigen, several have been previously linked to invasion in vertebrate models. Specifically, they identify the sulfhydryl oxidase Qsox1, Dtg and Put which relate to BMP-signalling, Dpp and Gp150 which modulates Notch signaling.
Can the characterization of Minerva be informative to vertebrate models? In other words, is Minerva activity conserved from flies to vertebrate? Sequence alignment revealed that the murine MFSD1 shares high sequence similarities with Minerva with 68% of amino acids conservation. MFSD1 shows the same localization patter in cells, colocalizing with markers for the Golgi. Most importantly, overexpression of MFSD1 in minerva mutant macrophages rescues O-glycosylation levels and the germband infiltration defects described, suggesting that the ability of Minerva to support tissue invasion is conserved up to mammals.
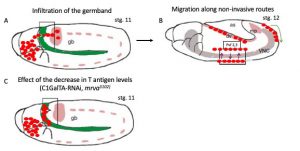
Figure 1: Schematic representation of the development routes Drosophilahemocytes follow to disperse throughout the embryo. A) Drosophilahemocytes (in red) originate in the head mesoderm and they populate the entire embryo by moving along 3 main routes. In the first route, hemocytes move over the yolk (in green) towards the germband (gb) and invade the epithelium. B) hemocytes then move along the developing ventral nerve cord (VNC) and through the dorso-ventral channels that run across it (purple arrows) engulfing and digesting apoptotic corpse and helping to shape the VNC. C) Reduction in the levels of O-glycosylation specifically in hemocytes alter their ability to infiltrate the germband causing an abnormal accumulation of cells sitting on the yolk.
Original picture was adapted from(Evans and Wood 2014). In the picture, dorsal is at the top and anterior to the left.
Relevance:
The work reported in this preprint contributes to the field of O-glycosylation providing two advances:
- It describes for the first time the O-glycoproteome of Drosophila
- It identifies a predicted MFS protein, Minerva, as a novel regulator of O-glycosylation and provides a genetically tractable model to address to what extent O-glycosylation influences cell migration, most specifically cell infiltration into neighbouring tissues.
The observation that this conserved protein regulates cell invasion through glycosylation offers a unique opportunity to investigate how cells use this PTM to bypass tissue barriers and penetrate into tissues, with important implications for cancer biology. Altered glycosylation is in fact an old dogma in cancer biology and often high levels of GalNAc-transferases have been linked to poor patient prognosis.
Questions to the authors:
- Very recently a model to study immune cell extravasation has been established in flies (Thuma et al. 2018). Do you consider turning on to this model to question whether O-glycosylation is involved in extravasation and to establish whether any parallelism can be drawn with the mechanism(s) of cancer cell extravasation?
- In your proteomic study, have you found any adhesion protein or protease whose levels of glycosylation are altered in minervamutants?
References:
Evans, I. R., and W. Wood. 2014. ‘Drosophila blood cell chemotaxis’, Curr Opin Cell Biol, 30: 1-8.
Thuma, L., D. Carter, H. Weavers, and P. Martin. 2018. ‘Drosophila immune cells extravasate from vessels to wounds using Tre1 GPCR and Rho signaling’, J Cell Biol, 217: 3045-56.
doi: https://doi.org/10.1242/prelights.7727
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cancer biology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Taxane-Induced Conformational Changes in the Microtubule Lattice Activate GEF-H1-Dependent RhoA Signaling
Vibha SINGH
ROCK2 inhibition has a dual role in reducing ECM remodelling and cell growth, while impairing migration and invasion
Sharvari Pitke
preLists in the cancer biology category:
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (No Ratings Yet)
(No Ratings Yet)