A high-coverage genome from a 200,000-year-old Denisovan
Posted on: 2 December 2025 , updated on: 7 December 2025
Preprint posted on 20 October 2025
A global map for introgressed structural variation and selection in humans
Posted on: , updated on: 7 December 2025
Preprint posted on 24 June 2025
Ancient Denisovan DNA from a 200,000-year-old genome and a new global map of archaic structural variants show that inherited Denisovan segments, including whole centromeres, still shape human diversity and local adaptation today
Selected by Siddharth SinghCategories: evolutionary biology, genetics, genomics, paleontology
A new high-coverage Denisovan genome, approximately 200,000 years old, and a global map of archaic structural variants reveal that repeated mixing with Denisovans has left DNA from single genes to whole centromeres in our genomes, and that some of these inherited pieces have contributed to human adaptation to their environments.
Why I highlight these preprints
Archaic introgression, the DNA that modern humans inherited from Neandertals and Denisovans, is known to comprise ~2-5% of Eurasian genomes. Yet, much of what we know has focused on single-nucleotide changes. These two preprints tackle the problem from new directions.
The preprint from Peyrégne and team presents a 200,000-year-old Denisovan genome, only the second high-quality Denisovan ever sequenced, shedding light on Denisovan population history and previously unrecognized gene flow events. The preprint by Hsieh and colleagues uses long-read sequencing and assembly to map archaic structural variants in modern humans, revealing that large segments of DNA (including entire centromeres) introgressed from Denisovans/Neandertals and may have been favored by natural selection.
I highlight both these studies because they push the boundaries of ancient DNA and pangenome research, and when combined, show how deeply archaic lineages and structural variation have influenced human adaptation in ways we’re only now beginning to understand.
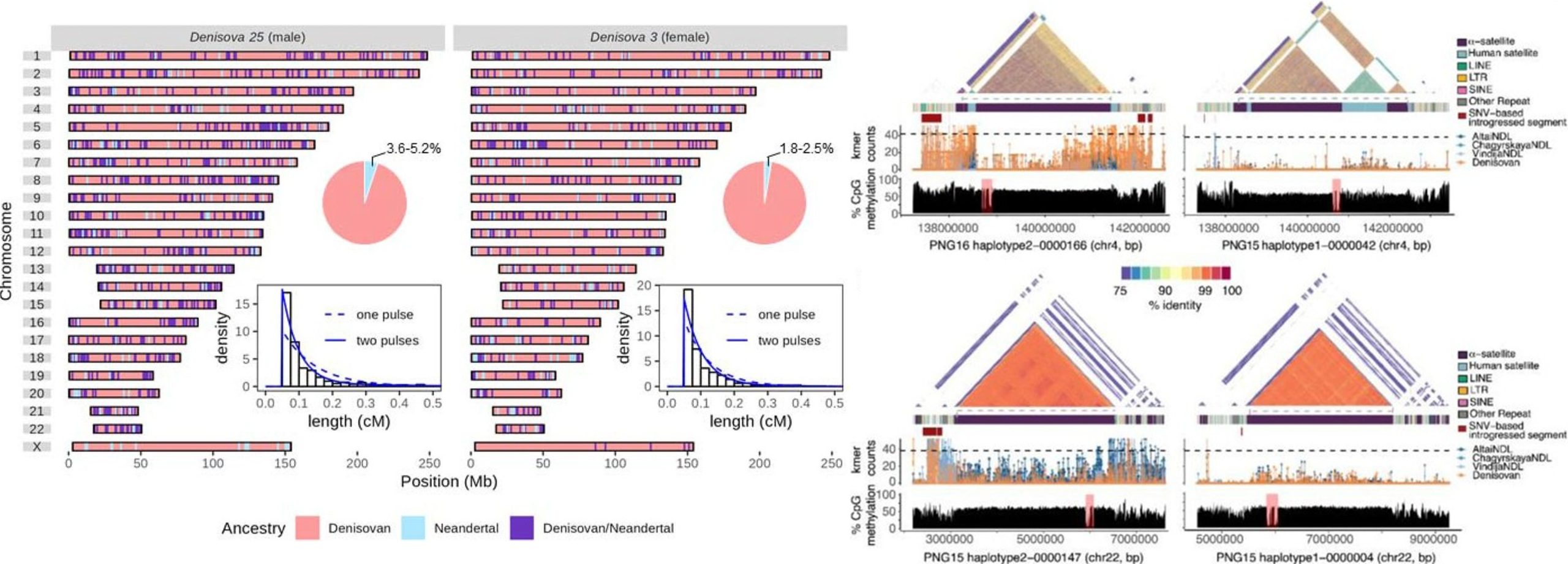
Second high-coverage Denisovan genome: Peyrégne and team sequenced a Denisovan male (“Denisova 25”) at ~23.6× coverage from a molar dated ~200,000 years before present (BP). His genome reveals a distinct, small Denisovan population that interbred with early Neandertals and was later replaced by Neandertals-admixed Denisovans. Remarkably, Denisova 25 also carries genetic material from an even more divergent hominin lineage (aka “super-archaic” humans) that split off before Denisovans and modern humans diverged. By comparing this genome to the younger Denisova 3 genome (~65k BP), the authors show that at least three distinct Denisovan lineages contributed DNA to present-day humans.
Global archaic SV map: Hsieh and colleagues built an unprecedented map of introgressed structural variation by integrating 4 new high-quality Papuan genomes with 94 diverse human assemblies. They discovered ~3,600 archaic introgressed SVs (≥50 bp), identified by finding SVs embedded in segments of Neanderthal/Denisovan origin. Notably, 44% of these introgressed SVs are located within genes (1,592 SVs), often in medically or functionally important regions. Using population-genomic scans (PBS statistics, coalescent modeling), the study finds 16 introgressed SVs with signatures of positive selection in Papuans. Many of these adaptive SVs involve immune-related genes and are associated with changes in gene expression in Papua New Guinea populations.
Cross-study mechanistic links
Ancient DNA meets pangenomics: These studies address a key challenge: ancient genomes are rich in information but lack long-range structure due to degradation, whereas modern long-read assemblies capture structural variants but require ancient references to recognize their origin. The Denisova 25 genome provides a new archaic reference point, while the SV map leverages modern Papuan genomes to unveil archaic SVs that short-read ancient DNA could not detect.
Together, they demonstrate a synergistic strategy, utilizing high-quality ancient sequences to inform introgression mapping and employing advanced modern assemblies to discover large archaic variants, thereby providing a more comprehensive picture of introgressed diversity than either approach alone.
Oceania as an archaic hotspot: Both preprints converge on Papua New Guinea/Oceania as a focal point of archaic influence. Peyrégne and team show that Oceanians carry a deeply divergent Denisovan lineage (a unique Denisovan ancestry not found in East Asians), consistent with an early southerly admixture event. Hsieh and colleagues accordingly find that Papuans harbor the highest concentration of introgressed SVs among global populations. In fact, many of the 16 adaptive SVs are specific to Papuans, and introgressed centromeres were identified in Papuan genomes but not in other groups.
This alignment suggests that Oceanians not only have the most Denisovan DNA by quantity, but also qualitatively different archaic variants, likely stemming from that extra Denisovan lineage, which have been retained and favored in these populations.
Adaptive legacy of archaic DNA: A theme across both studies is that archaic admixture wasn’t just a curiosity; it contributed functionally significant variation that persists today. The Denisova 25 preprint identifies multiple introgressed segments that reached high frequency in specific regions, suggesting local adaptive benefits (they identified 38 candidate Denisovan introgressed loci under selection in various populations). The SV preprint directly pinpoints structural variants under selection. Both results underscore that introgressed DNA was not purely deleterious noise diluted over time; instead, specific archaic alleles, whether small nucleotide changes or large SVs, conferred adaptive advantages (immunity, metabolism, perhaps altitude or pathogen resistance) that helped humans thrive in new environments.
This shared finding bolsters the view that admixture with archaic humans provided a creative genetic source for adaptation, enriching our species’ evolutionary toolkit.
Field impact and following experiments
Richer archaic genealogies: Having a second Denisovan genome ~135,000 years older than the first allows researchers to reconstruct a much more detailed Denisovan family tree. This will spur new models of Pleistocene human dispersal and underscore that the “Denisovan” was not a single, homogeneous group.
Beyond SNPs – Introgression of Large Variants: The SV-focused preprint by Hsieh and colleagues expands the scope of introgression research from single-point mutations to structural genome archaeology. It reveals that entire regions, such as centromeres and megabase-scale inversions, can be introgressed and survive in our gene pool. This opens up new questions about genome stability and incompatibility.
Methodological leap for detecting ancient DNA: The integration of long-read genome assemblies from diverse living individuals, as demonstrated here, provides a powerful tool for capturing introgression that traditional short-read analyses often miss. This pangenome approach can be extended: researchers can apply similar pipelines to African populations (to hunt for “ghost” archaic SVs from unknown hominins) or other underrepresented groups. The success of identifying adaptive SVs also encourages the development of new statistics and machine-learning methods tuned for structural introgression signals, analogous to those long used for SNPs.
Functional follow-ups on adaptive introgressions: Both preprints yield concrete targets for experimental biology. We now have specific Denisovan-derived alleles, from a FOXP2 regulatory change to a 55 kb HLA deletion, that can be functionally tested for their effects on gene expression or organismal phenotypes. This paves the way for mechanistic studies of archaic alleles, using CRISPR to recreate archaic variants in cell lines or mice to observe immune responses, or epidemiological studies in humans to see if carriers of certain introgressed SVs have different health outcomes.
The identification of archaic centromeres particularly raises the hypothesis that they might impact fertility or meiotic segregation, a testable idea in model systems. By highlighting these variants, the work generates new hypotheses about how archaic genetics contributed to traits such as immunity, high-altitude adaptation, and even morphology, shifting the field toward a more integrative understanding of human adaptation that links deep evolutionary history with present-day biology.
Author questions
Peyrégne et al. 2025
- Super-archaic admixture: The Denisova 25 analysis reports tentative gene flow from a very ancient hominin lineage, with a high false-positive rate for candidate segments. What specific additional genomes or statistical tests would most clearly confirm or refute super-archaic DNA in Denisovans?
- Deep Denisovan lineage in modern humans: You infer that Oceanians and South Asians carry DNA from a deeply divergent Denisovan population likely isolated in South Asia that is not shared with East Asians. Which specific markers or haplotypes best distinguish this southern Denisovan ancestry from the Denisova 3-like northern ancestry, and how could future Denisovan or South Asian ancient genomes most decisively test this separate southern lineage model?
- Adaptive introgression targets: Your Denisova 25 scan identifies candidate selected segments near ATP6V0A4 and COL4A3 in South Asians that also carry disease-linked annotations. Which concrete functional and population-level analyses do you see as most informative for testing whether these Denisovan-derived variants are genuinely adaptive rather than hitchhiking or mildly deleterious?
Hsieh et al. 2025
- Archaic centromeres and fitness: You report intact archaic-derived centromeres in Papuan genomes with divergence times that match Neandertal and Denisovan splits. What empirical tests or datasets would most directly demonstrate whether these introgressed centromeres affect recombination fertility or missegregation risk, and whether they represent neutral variants, tolerated variants, or targets of selection?
- SV introgression map: Your map is built from phased assemblies for only 49 individuals, which underrepresent Africa, the Americas, and parts of Asia. What are your concrete plans to extend long-read assemblies to these populations and to search for additional introgressed SVs, including possible ghost archaic SVs in sub-Saharan Africa, and how general do you expect the Papuan-based signals of archaic SV adaptation to be once a broader panel is available?
- Integrating Denisova 25 with SV data: Now that the Denisova 25 genome is available, do you plan to cross-reference it with the introgressed SV catalog? Denisova 25 might contain structural variants that were not present in Denisova 3. Have you checked if any of the 16 adaptive SVs identified in Papuans are actually present in Denisova 25’s assembly?
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
Also in the genetics category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Also in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the paleontology category:
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Evaluating the impact and detectability of mass extinctions on total-evidence dating
Tom Carruthers
Classification of dinosaur footprints using machine learning
Ryan Harrison
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the genetics category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |











 (No Ratings Yet)
(No Ratings Yet)