Combining antibiotics with antivirulence compounds is effective and can reverse selection for antibiotic resistance in Pseudomonas aeruginosa
Posted on: 10 December 2019 , updated on: 12 December 2019
Preprint posted on 5 December 2019
Article now published in PLOS Biology at http://dx.doi.org/10.1371/journal.pbio.3000805
No use resisting: reversing antibiotic resistance using combinations of antibiotics and anti-virulence compounds
Selected by Zhang-He GohCategories: microbiology, pharmacology and toxicology
Background of preprint
ESKAPE—Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter—is a group of six pathogens exhibiting multidrug resistance and virulence that was first described in 2008 [1]. Today, these six pathogens continue to present a major concern to the World Health Organisation (WHO), which has characterised them as “pathogens for which antibiotics are urgently needed” [2,3]. Specifically, carbapenem-resistant P. aeruginosa has been listed in a high priority group [2], testament to the increasing urgency with which this problem must be resolved.
In their preprint, Rezzoagli et al. focus on P. aeruginosa for its clinical relevance and its position as a model organism for anti-virulence research. The authors investigate two major anti-virulence mechanisms: quorum sensing and siderophore-mediated iron-uptake systems, using furanone c-30 and gallium respectively (Fig. 1A). These compounds were investigated in combination with existing antibiotics (Fig. 1B) used to treat P. aeruginosa in four major steps (Fig. 2).
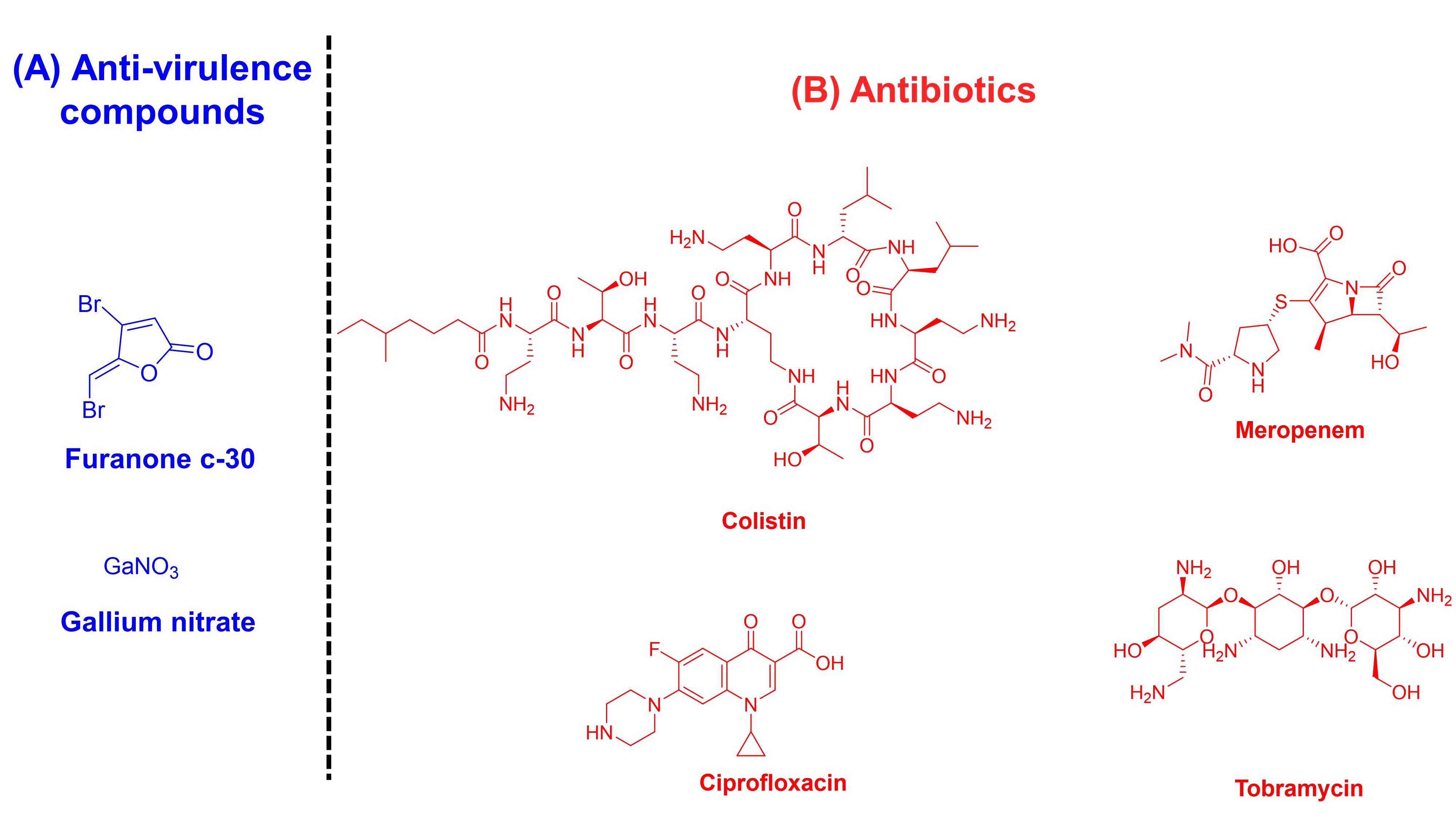
Fig. 1. Structures of compounds investigated in this preprint, categorised into their respective mechanisms of action. (A) Anti-virulence compounds: furanone c-30 and gallium nitrate. (B) Antibiotics: ciproflxacin, meropenem, colistin, and tobramycin.
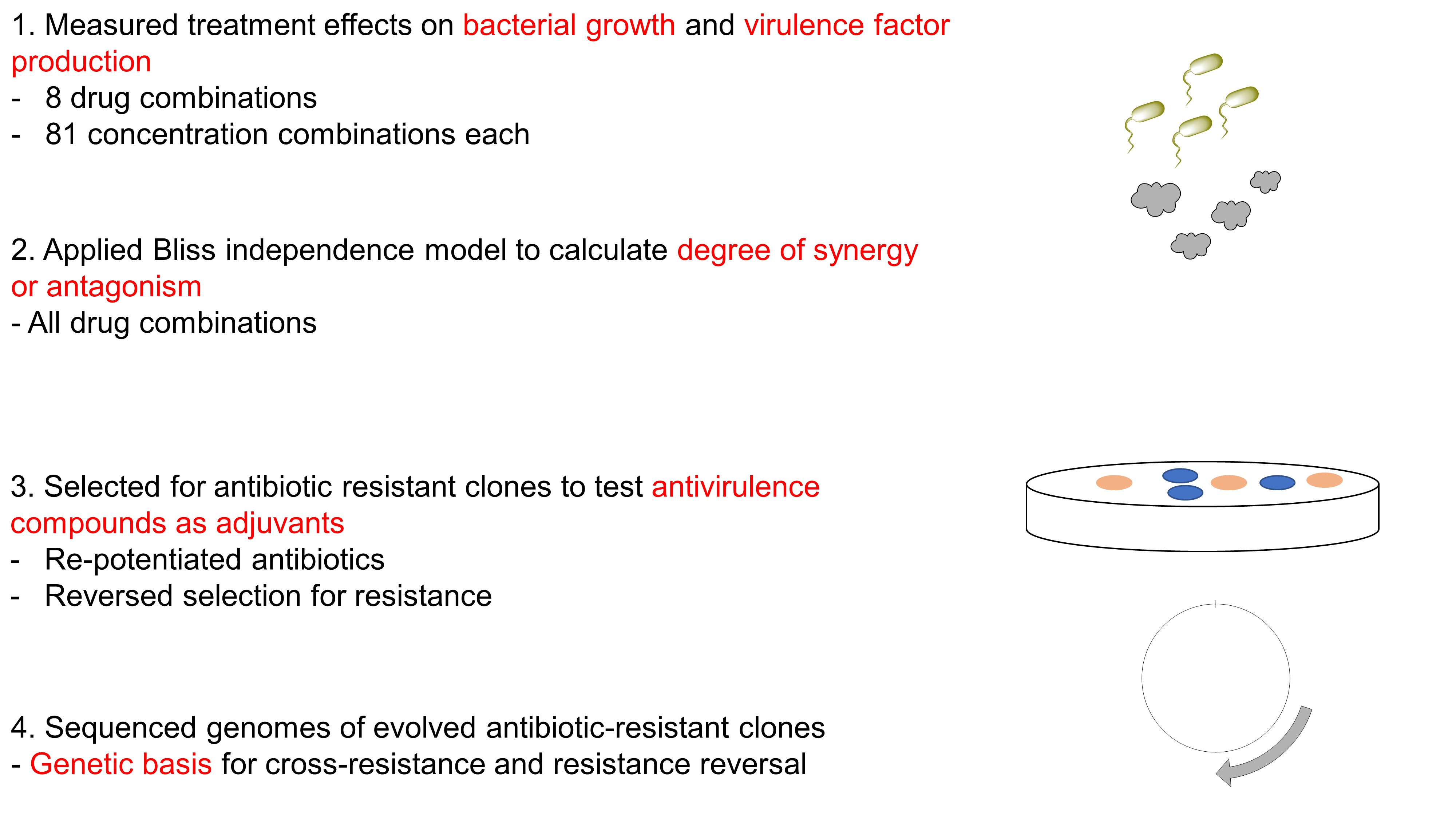
Fig. 2. Four steps undertaken in this preprint.
Key findings of preprint
(A) Treatment effects on bacterial growth and virulence factor production
The authors determined that the dose-response curve to the four antibiotics and the two anti-virulence compounds were sigmoidal (preprint Figure 1). The antibiotic concentrations could be classified into three ranges (Table 1).
Table 1. Three concentration ranges of antibiotics.

The anti-virulence compounds were tested in both normal and enriched media—the latter of which did not require virulence factors for growth. This led the authors to conclude that there is a window of concentrations for which growth inhibition is solely attributed to virulence factor quenching, and that high concentrations of anti-virulence compounds may curb growth.
(B) Degree of synergy or antagonism between anti-virulence compounds and antibiotics
Rezzoagli et al. investigated different gallium- and furanone c-30-antibiotic combinations (Table 2, Table 3), and examined whether the degrees of synergy correlated between growth and virulence factor inhibition (Table 4). Based on their observations, the authors concluded that specific drug combinations cannot be classified as either synergistic or antagonistic; rather, they emphasise the importance of separately characterising these interactions stratified by concentration, with the strongest effects observed at intermediate drug concentrations.
Table 2. Effect of combination on bacterial growth
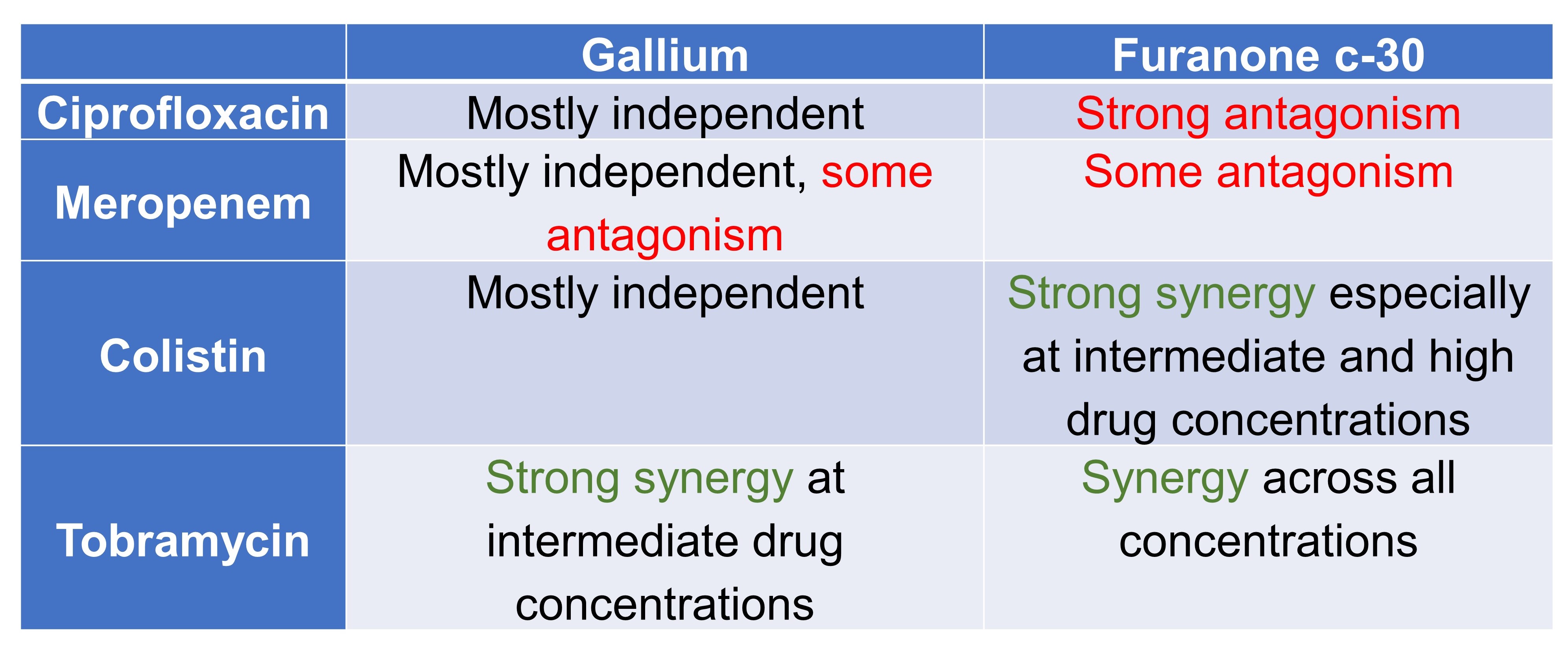
Table 3. Effect of combination on virulence factor production
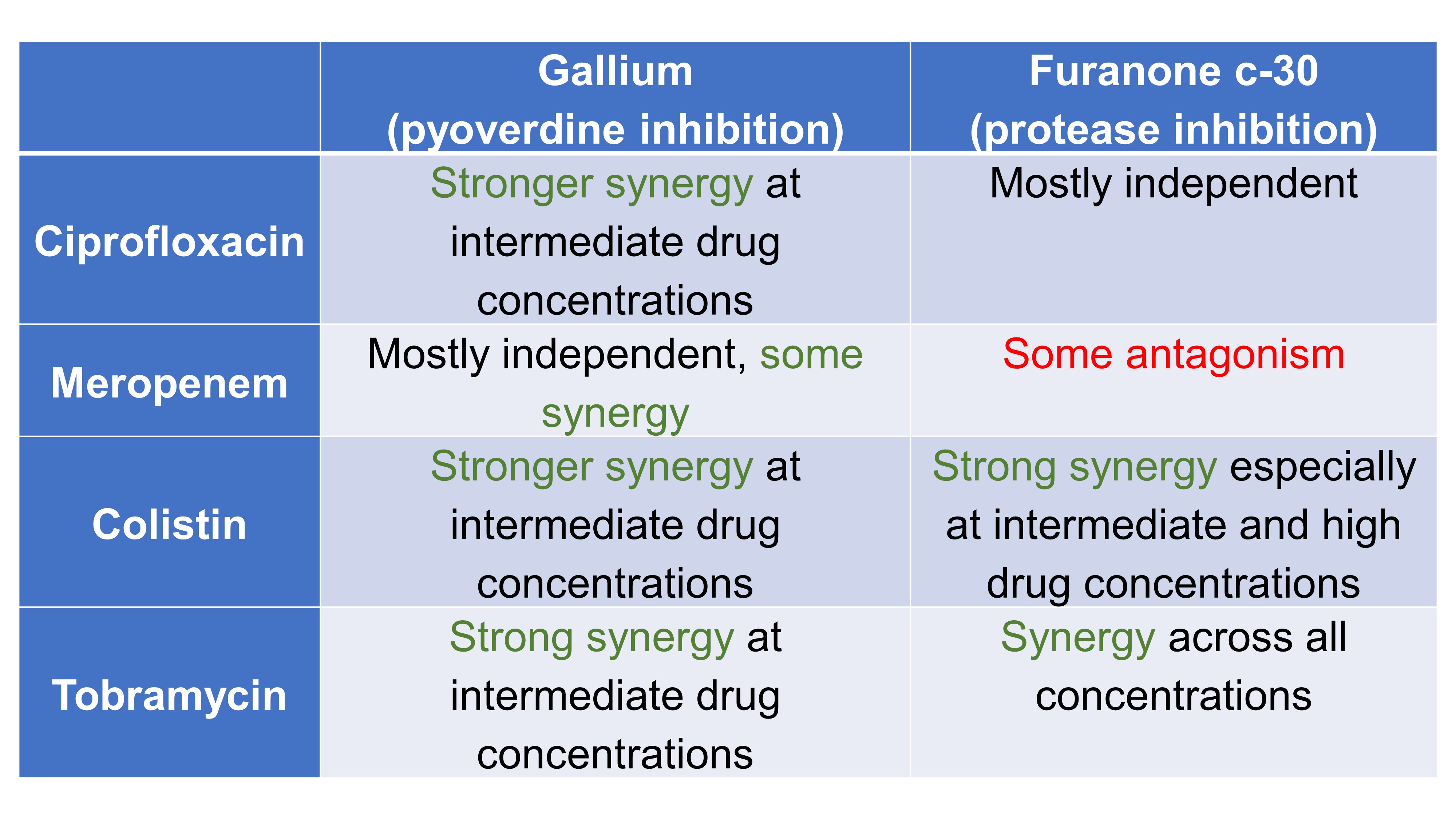
Table 4. Association among degrees of synergy between growth and virulence factor inhibition.
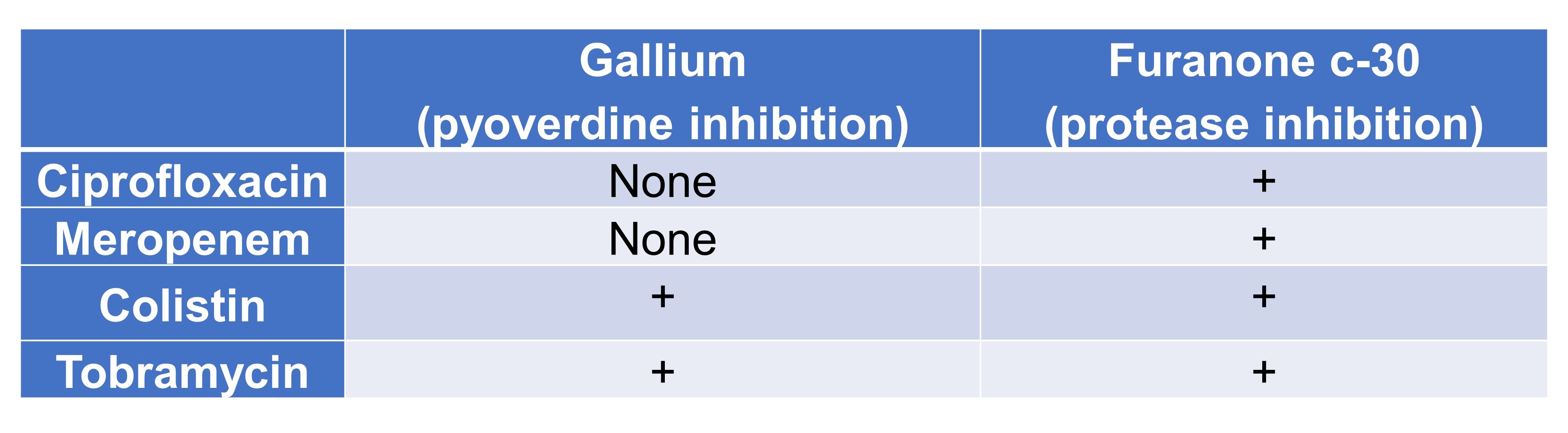
(C) Antivirulence compounds as adjuvants
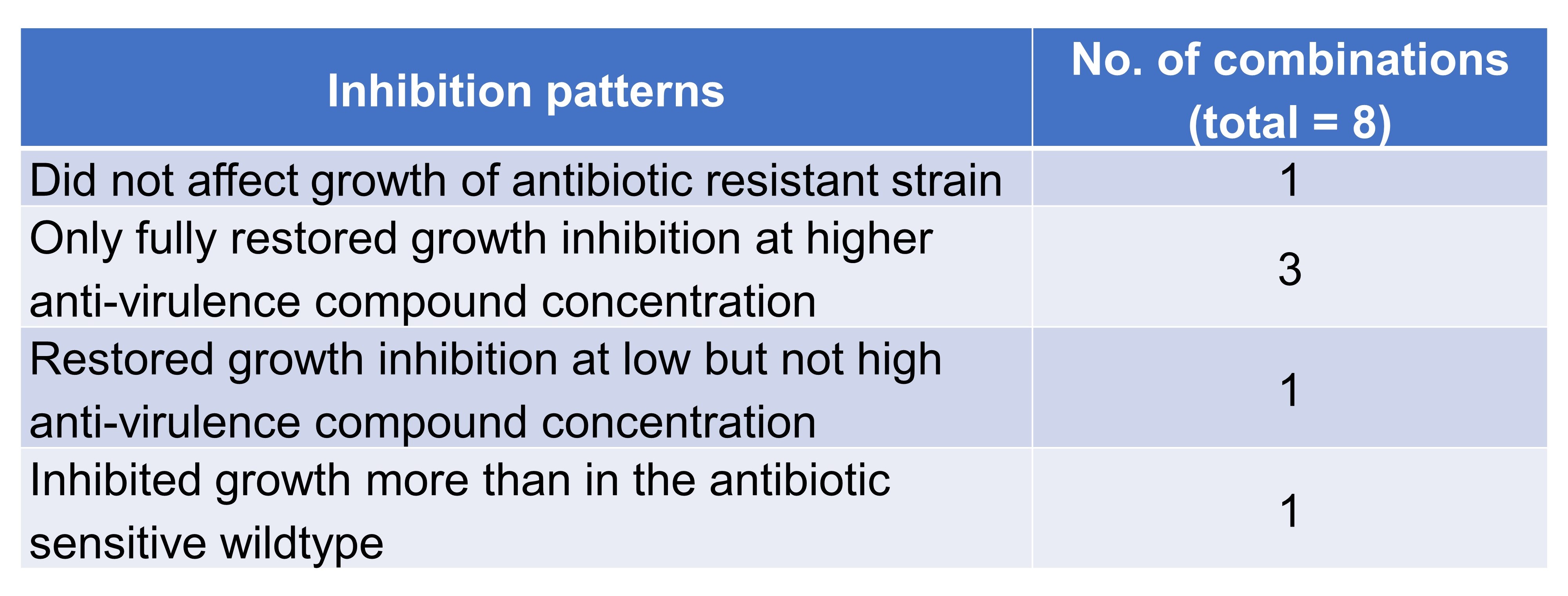
Rezzoagli et al. then found that, when anti-virulence compounds and antibiotics were used in combination, growth inhibition of resistant clones was restored in seven out of eight drug combinations, with four recurring inhibition patterns (Table 5). Using competition assays, the authors showed that the addition of anti-virulence compounds had variable and combination-specific effects on the spread of antibiotic-resistant (AtbR) clones.
Table 5. Inhibition patterns exhibited by combinations of anti-virulence compounds and antibiotics.
(D) Genetic basis for antibiotic resistance
Finally, the authors conducted whole-genome sequencing on their antibiotic-resistant clones, finding single nucleotide polymorphisms (SNP) and insertion or deletion of bases (INDEL) that have been associated with resistance to the respective antibiotics (preprint Table 1).
What I like about this preprint
As the threat of antimicrobial resistance looms, pharmaceutical researchers, healthcare professionals, and policymakers are scrambling to find ways to reduce our reliance on antibiotic use. Consequently, alternative strategies to tackle the problem are returning—researchers are beginning to explore the use of bacteriophages and anti-virulence compounds as alternatives to conventional antibiotics [4,5]. This preprint, in which Rezzoagli et al. utilise P. aeruginosa as a model organism in their investigation of the interaction between anti-virulence compounds and antibiotics, is thus a timely undertaking.
Future directions
Rezzoagli et al. proposed three areas of further research in their preprint: to quantify the rate of resistance evolution under the combinatorial treatments, to test the combinations in relevant animal-host models, and to better characterise the combinations’ pharmacodynamics and pharmacokinetics. Indeed, the highly exploratory nature of this work means that anti-virulence compounds will need to be further evaluated before it can complete its journey to the clinic.
These three areas are intricately linked. The rate of resistance evolution in vitro and in vivo may not correlate, and investigations pertaining to this topic would depend on the animal-host model used. Pharmacokinetics vary across animals, so the animal-host model will have to be carefully selected for the pharmacokinetics—and therefore the pharmacodynamics—of these combinations to be representative of that in humans. In fact, this will be a particularly challenging but important undertaking, given the varying correlations between synergy and antagonism of the combinations on growth and virulence factor inhibition.
Many questions remain. How should the evolution of resistance be characterised, and if possible, modelled? How applicable are these findings in P. aeruginosa to other pathogens, especially ESKAPE? What animal-host models should be used to represent the pharmacodynamics of infection and the pharmacokinetics of the drug combinations? What are the potential toxicities of these drug combinations? How would the pharmacokinetics-pharmacodynamic effect play out clinically in patients suffering from kidney and liver impairment? Presumably, these combinations will be used as a drug of last resort in patients who are very ill, so the functions of the major metabolising (liver) and excreting (kidney) organs need to be taken into consideration.
Near the end of the eponymous musical, the Phantom of the Opera declared triumphantly that “the games are at an end”. Our misuse and overuse of antibiotics have pushed us to the point of no return, and the parallels to the equally spooky spectre of running out of effective antibiotics are painfully clear. In finding new strategies in our uphill battle, I hope that we will one day—like the Phantom—proclaim that there is “no use resisting”.
References
[1] Rice LB, Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE, The Journal of Infectious Diseases 197(8) (2008) 1079-1081.
[2] Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Aboderin AO, Al-Abri SS, Awang Jalil N, Benzonana N, Bhattacharya S, Brink AJ, Burkert FR, Cars O, Cornaglia G, Dyar OJ, Friedrich AW, Gales AC, Gandra S, Giske CG, Goff DA, Goossens H, Gottlieb T, Guzman Blanco M, Hryniewicz W, Kattula D, Jinks T, Kanj SS, Kerr L, Kieny M-P, Kim YS, Kozlov RS, Labarca J, Laxminarayan R, Leder K, Leibovici L, Levy-Hara G, Littman J, Malhotra-Kumar S, Manchanda V, Moja L, Ndoye B, Pan A, Paterson DL, Paul M, Qiu H, Ramon-Pardo P, Rodríguez-Baño J, Sanguinetti M, Sengupta S, Sharland M, Si-Mehand M, Silver LL, Song W, Steinbakk M, Thomsen J, Thwaites GE, van der Meer JWM, Van Kinh N, Vega S, Villegas MV, Wechsler-Fördös A, Wertheim HFL, Wesangula E, Woodford N, Yilmaz FO, Zorzet A, Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis, The Lancet Infectious Diseases 18(3) (2018) 318-327.
[3] Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR, Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review, Frontiers in microbiology 10 (2019) 539-539.
[4] Fleitas Martínez O, Cardoso MH, Ribeiro SM, Franco OL, Recent Advances in Anti-virulence Therapeutic Strategies With a Focus on Dismantling Bacterial Membrane Microdomains, Toxin Neutralization, Quorum-Sensing Interference and Biofilm Inhibition, Front Cell Infect Microbiol 9 (2019) 74-74.
[5] Theuretzbacher U, Outterson K, Engel A, Karlén A, The global preclinical antibacterial pipeline, Nature Reviews Microbiology (2019).
doi: https://doi.org/10.1242/prelights.15677
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Characterization of natural product inhibitors of quorum sensing in Pseudomonas aeruginosa reveals competitive inhibition of RhlR by ortho-vanillin
UofA IMB565 et al.
Feedback loop regulation between viperin and viral hemorrhagic septicemia virus through competing protein degradation pathways
UofA IMB565 et al.
Lytic bacteriophages interact with respiratory epithelial cells and induce the secretion of antiviral and proinflammatory cytokines
UofA IMB565 et al.
Also in the pharmacology and toxicology category:
G6b-B antibody-based cis-acting platelet receptor inhibitors (CAPRIs) as a new family of anti-thrombotic therapeutics
Simon Cleary
Pervasive sublethal effects of agrochemicals as contributing factors to insect decline
Roberto Amadio
Mixed Alkyl/Aryl Phosphonates Identify Metabolic Serine Hydrolases as Antimalarial Targets
Zhang-He Goh
preLists in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the pharmacology and toxicology category:
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Drug use in special populations
Any drugs that are being used in special populations: Patients with liver and kidney failure, in paediatrics, in geriatrics, and in pregnant or lactating patients. Includes the discovery of factors that could potentially affect drug use in these special populations.
| List by | Zhang-He Goh |
Toxicology of toxicants, existing therapeutics, and investigational drugs
Preprints that describe the toxicology of environmental pollutants and existing and upcoming drugs. Includes both toxicokinetics and toxicodynamics, as well as technological improvements that will help in the characterisation of this field.
| List by | Zhang-He Goh |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)