Downstream of gasdermin D cleavage, a Ragulator-Rag-mTORC1 pathway promotes pore formation and pyroptosis
Posted on: 9 December 2020
Preprint posted on 2 November 2020
Article now published in Cell at http://dx.doi.org/10.1016/j.cell.2021.06.028
Categories: immunology, systems biology
Background:
Pyroptosis is an inflammatory form of cell death characterized by membrane disruption leading to lytic cell death and release of inflammatory molecules, including IL-1 family cytokines. This membrane disruption is the result of pores formed by the protein gasdermin D (GSDMD), which is cleaved by inflammatory caspases following infection or inflammasome activation. The N-terminal fragment of cleaved GSDMD (NT-GSDMD) localizes to the membrane and oligomerizes to form pores which permit the transit of relatively small molecules (including IL-1 family cytokines) and eventually lead to full lytic cell death. While the upstream events leading to GSDMD cleavage have been more deeply studied, the extent of regulation of pore formation remains unclear. In this preprint, Evavold et al present a forward genetic screen and identify a novel regulatory mechanism of GSDMD pore formation.
Key findings:
- Development of a system to study GSDMD pore formation
A key feature of this preprint is the development of a biological system that allows the specific study of the pore formation stage of pyroptosis. In order to do this, the authors established a cell line system expressing a doxycycline-inducible NT-GSDMD-BFP fusion protein. Induction of NT-GSDMD-BFP expression is sufficient to cause pore formation independent of the upstream pyroptosis machinery (e.g. inflammasome activation), which is easily monitored by uptake of the dye propidium iodide (PI). BFP fusion allows for tracking of NT-GSDMD expression and localization, which is critical to deconvolute the steps of membrane localization and subsequent pore formation. These cell lines were developed in a background of Cas9 transgenic expression, allowing for CRISPR knockout of regulators of the pore formation process. A schematic overview of this system is presented in Figure 1. - Ragulator/mTORC1 regulate GSDMD pore formation
The authors used their new system to perform a genome-wide CRISPR screen to identify regulators of pore formation. By examining guide sequences depleted from the highest BFP+/PI- cells, the authors identified genes specifically required for pore formation downstream of GSDMD cleavage. The top hits included multiple components of the Rag-Ragulator complex, which is responsible for recruiting mTORC1 to lysosomes for amino acid sensing. Follow-up experiments using targeted knockouts revealed that Ragulator and mTORC1, but not mTORC2, are required for proper pore formation and pyroptosis, but not for membrane localization of NT-GSDMD., In natural pyroptotic settings, such as Salmonella infection, mTORC1-deficient cells were resistant to pyroptosis, even while they showed proper GSDMD cleavage, confirming the role of Ragulator/mTORC1 in regulating GSDMD pore formation.
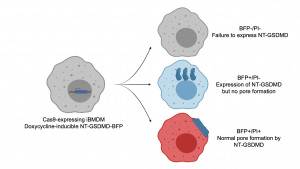
Figure 1: Schematic of system to study GSDMD pore formation
An immortalized BMDM cell line expressing Cas9 and a doxycycline-inducible NT-GSDMD-BFP fusion protein was generated. After induction with doxycycline, several outcomes are possible. Failure to properly express the NT-GSDMD fusion protein will result in a BFP and PI-negative cell (propidium iodide, taken up through pores in the membrane). Expression of the NT-GSDMD-BFP fusion protein without pore formation will result in a BFP single-positive cell. Proper pore formation as in normal pyroptosis will result in PI uptake and a BFP/PI double-positive cell. Figure prepared with biorender.
Importance:
This study decouples GSDMD cleavage and membrane recruitment from pore formation and downstream cytokine release and cell death. This adds a new layer of regulatory potential to the process of pyroptosis, and promises to reshape how we understand this process of regulated cell death. The potential integration of cellular metabolic state or activity with GSDMD pore formation, through Ragulator/mTORC1, also presents an intriguing new area of study in the field of cell death.
Moving forward / Questions for authors:
- The mechanistic process by which Ragulator/mTORC1 regulates pore assembly will be exciting to unravel in future studies. One straightforward question is to ask whether Ragulator/mTORC1 activity is required *during* the process of pyroptosis. Pharmacologic inhibitors could help answer the question of whether Ragulator/mTORC1 activity is required during/after membrane recruitment of NT-GSDMD to facilitate pore formation, or if their deficiency “primes” the cell in advance to be susceptible to pore formation. Either result would be interesting, suggesting either that certain metabolic activities are a key part of active pyroptosis or that certain cells are particularly susceptible/resistant to pyroptosis based on their metabolic state prior to infection.
- What do the depleted sgRNAs from the screen (i.e. the opposite end of the screen from the Ragulator components, shown in Fig 3A) represent? Does the screen distinguish between potential inability to express/maintain the NT-GSDMD fragment (BFP- cells) and increased cell death (BFP+/PI+ cells)?
doi: https://doi.org/10.1242/prelights.26212
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the immunology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Scalable transcription factor mapping uncovers the regulatory dynamics of natural and synthetic transcription factors in human T cell states
Inês Caiado
Also in the systems biology category:
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Longitudinal single cell RNA-sequencing reveals evolution of micro- and macro-states in chronic myeloid leukemia
Charis Qi
Environmental and Maternal Imprints on Infant Gut Metabolic Programming
Siddharth Singh
preLists in the immunology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the systems biology category:
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)