Evidence for an Integrated Gene Repression Mechanism based on mRNA Isoform Toggling in Human Cells
Posted on: 30 October 2018 , updated on: 31 October 2018
Preprint posted on 5 October 2018
Article now published in G3: Genes|Genomes|Genetics at http://dx.doi.org/10.1534/g3.118.200802
Categories: genetics
Background
Gene regulation is generally thought to be a linear process where transcriptional and translational regulation play independent roles in modulating gene expression. In the canonical model, mRNAs transcribed from a given gene always carry the same set of information and are later modified by other, independent mechanisms such as splicing or RNA editing. However, widespread alternative promoter usage, as shown by high-throughput transcription start site (TSS) mapping, suggests that mRNAs (from the same gene) generated by different TSSs can carry different information in their 5’ leaders, which in turn instructs translation. For example, expression can be altered by simply having one isoform contain an upstream open reading frame (uORF) that prevents proper translation of the correct ORF. The authors have first dissected this mechanism in the context of a single, essential gene and later have shown it to be pervasive in yeast meiosis. However, whether or not mammalian cells use such an integrated mechanism remains unknown. This preprint highlights a specific oncogene in humans, MDM2, where such a mechanism occurs and show that it might play an important role in regulating stem cell differentiation.
Key findings
The primary claim of the preprint is that MDM2 is regulated by an integrated transcriptional and translational mechanism. By toggling between two separate promoters, two mRNA transcripts with different translational efficiencies are produced (Figure 1). To test this hypothesis, the authors defined 3 key features of integrated regulation from their previous observations in yeast and proceeded to test whether these mechanisms also apply to MDM2 regulation.
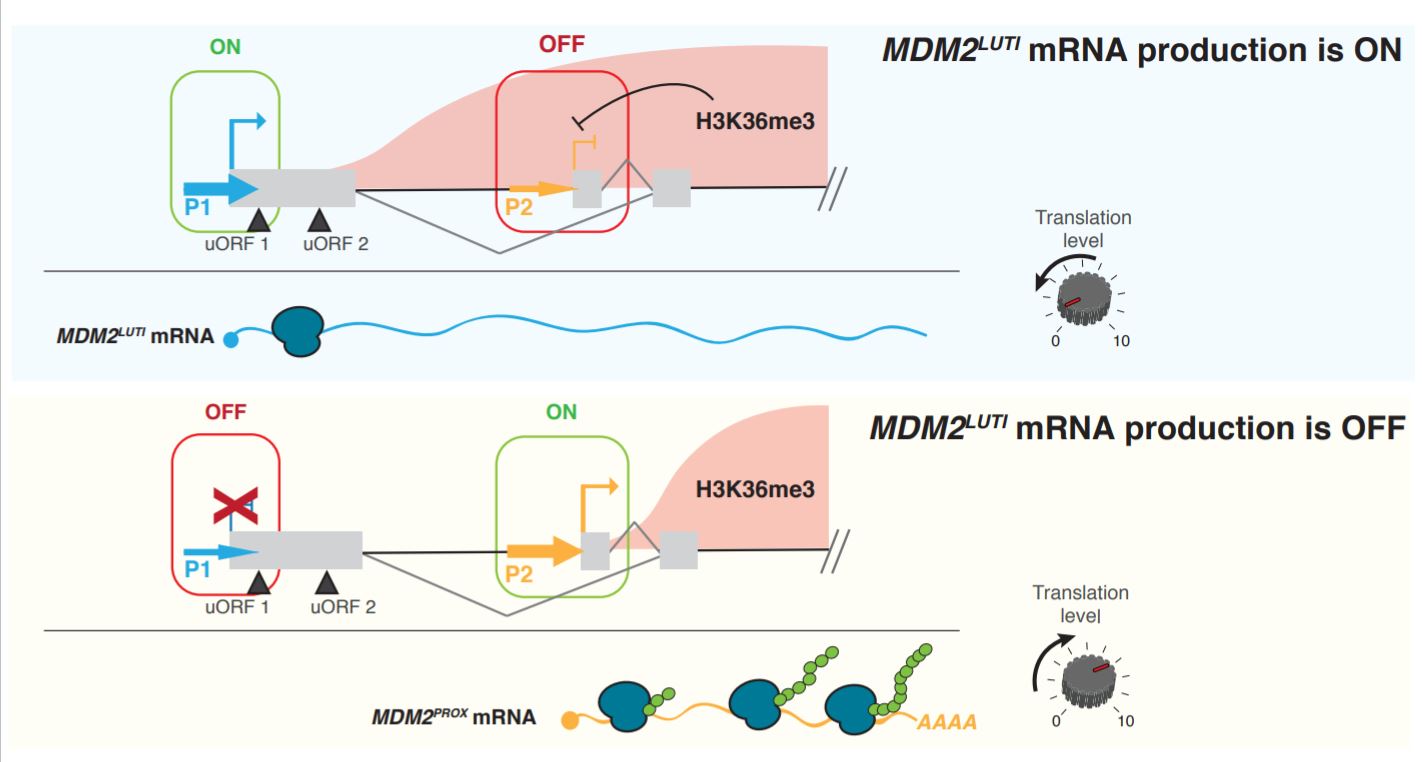
Figure 1. Summary of MDM2 regulation by an integrated mechanism (Figure 5 of the preprint).
One, the isoform from the distal promoter contains an uORF that represses proper translation. MDM2 has previously been shown to express two different isoforms, a longer isoform called MDM2LUTI and a shorter isoform, MDM2PROX, from the distal (P1) and proximal (P2) promoters respectively. MDM2LUTI contains 2 additional uORFs in its 5’ leader, and is therefore translated less efficiently than MDM2PROX, which only carries the main ORF (and no uORF).
Two, transcription from the distal promoter disrupts transcription from the proximal promoter, allowing for a complete switch of promoter usage. This step is extremely crucial to ensure that only the desired transcript is produced. To test this, the authors used a CRISPR/dCas9-based transcription inhibition technique using a catalytically dead Cas9 (CRISPRi) to inhibit expression at the P1 promoter, and showed that expression of MDM2PROX was indeed increased. The same trend was observed despite inhibition of p53 expression, a known activator of the P2 promoter, suggesting that the upregulation of MDM2PROX was independent of p53 activation. These results imply that P1 most likely inhibits P2 expression directly through transcriptional inhibition rather than some secondary pathway. To understand how this inhibition is achieved, the authors then measured the levels of H3K36me3 in the promoters and gene body of MDM2. H3K36me3 is a co-transcriptional modification, and is thus a mark of active transcription. It has also been implicated in repression of spurious transcription. Indeed, when P1 is inhibited, there is a decrease in H3K36me3 signal at the P1 and P2 promoters. This suggests that when P1 is transcribed, H3K36me3 is deposited at P2, which then represses it activity.
Finally, usage of the two promoters should be actively regulated by a developmental switch of some sort. In this preprint, the authors assessed this using an in vitro differentiation model, where they induced human embryonic stem cell (hESC) differentiation into neuronal and endodermal lineages and measured promoter usage throughout the differentiation process. They found that neuronal precursor cells and endodermal cells appear to prefer MDM2PROX, despite higher expression of MDM2LUTI than MDM2PROX in hESCs. This suggests that promoter usage for MDM2 is developmentally regulated, and the integrated regulation could represent a mechanism by which proper gene expression is achieved during development.
What I liked about the preprint
Gene regulation is generally thought of as a complex series of steps. However, the idea that transcriptional and translational regulation can be integrated actually simplifies the model. If the different steps are independent, we would have had to evolve mechanisms of controlling each step for optimal expression. Using this mechanism, the regulation is at the level of the promoter toggle, and translational regulation is simply a consequence of transcription. Personally, I find this idea quite comforting, because it means there are less steps to conquer in our understanding of gene expression. This mechanism also highlights how non-coding DNA can be used to regulate translation instead of transcription, which is something that we can use to understand the role of non-coding variants in development and disease. Finally, I think that the authors did a really good job laying out the hypothesis and its predictions and then systematically testing each prediction, something that is too often lacking in many scientific papers.
Future directions and questions
The obvious next step from this finding is to figure out how ubiquitous this regulation mechanism is in human cells, especially for developmentally important genes. I would also be very interested to understand exactly what signals determine which promoter is used, since it seems like the MDM2LUTI is preferred in neuronal precursors and endodermal cells, but not in differentiated neurons. Specifically, since the expression of MDM2 was only measured up to day 4 of endodermal differentiation, if they were allowed to grow further, would those cells also switch back to MDM2LUTI or continue to prefer MDM2PROX ?
Furthermore, H3K36me3 is postulated to be the mechanism by which the P1 transcript represses transcription from P2. It would be important to see what happens when the H3K36me3 writer enzyme in the cell types used is downregulated. Is P1 and P2 transcription now able to occur simultaneously, or does active transcription from P1 simply limit RNA polymerase access to P2 and thereby physically block transcription? Understanding how promoter usage is regulated could lead to many insights into basic transcriptional mechanisms and disease, such as the switch of the MDM2 promoter observed in cancer.
doi: https://doi.org/10.1242/prelights.5334
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genetics category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
preLists in the genetics category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)