Phenotypic landscape of schizophrenia-associated genes defines candidates and their shared functions
Posted on: 20 July 2018
Preprint posted on 3 July 2018
Article now published in Cell at http://dx.doi.org/10.1016/j.cell.2019.01.048
From GWAS to function: a phenotypic atlas of 132 zebrafish mutants sheds light on the role of human schizophrenia-associated genes
Selected by Daniel GrimesCategories: animal behavior and cognition, genetics, neuroscience
Background
Psychiatric disorders are a major healthcare concern, accounting for up to one quarter of worldwide disabilities. There are many challenges to understanding the biological basis of these diseases. First, psychiatric disorders are non-Mendelian; they involve many loci and inheritance patterns are therefore complicated. In 2014, a large genome-wide association study implicated over 100 loci in schizophrenia; each had a small effect. A second challenge is to understand the molecular mechanisms underpinning a psychiatric disorder, whether they be changes in neuronal activity or connectivity, changes in glial cell gene expression or countless other potential pathologies. The third, and perhaps most difficult, challenge is to understand how the molecular pathologies manifest in the context of the brain, the human organism, and outside environmental factors to control a biological output as multifaceted and nuanced as behavior.
An important step on the road to meeting these challenges is to define the roles of genes associated with psychiatric disorders in brain function by using animal models. The zebrafish, Danio rerio, is an exquisite system for this purpose. Brain anatomy, neural subtypes and aspects of development are conserved between humans and zebrafish, while behaviors such as anxiety and sociability can be assessed with high throughput. Imaging of brain activity is also possible in the semi-transparent larval stages. Importantly, most human disease genes are conserved in zebrafish and, now with the advent of CRISPR/Cas, the generation of mutant lines is increasingly tractable.
Overview of the work
Summer Thyme, Alexander Schier and colleagues have used all of these advantages to investigate the potential roles of genes located within schizophrenia-associated loci. In a heroic effort described in the current preprint, the team generated 132 mutant zebrafish lines and assessed them for behavioral defects, brain structural malformations, and perturbations to brain activity. While only a small number of mutants exhibited lethality prior to adulthood, more than half showed brain activity and/or behavioral phenotypes.
The researchers searched for behavioral phenotypes in larvae, kept in multi-well plates so that the experiment could be done with high-throughput, across a 2-day experiment (4-6 days post fertilization [dpf]) (Figure 1). Measures included how much the larvae moved and where they preferred to be in their dish, their sleep behavior and their responses to various stimuli (light flash, dark flash, acoustic startle, heat and others). Larvae were also tested for defects in habituation, the diminishing of a response to repeated stimuli, as well as prepulse inhibition, where a weak pre-stimulus reduces the reaction to a subsequent stronger stimulus. Across the library of mutants assessed, phenodeviants, mutants departing significantly from the wild-type, were discovered in all of these behavioral assays, and many mutants were affected in several tests. This revealed that different aspects of behavior can be impacted, and to different extents, by mutations in schizophrenia-associated genes, something which further underlines the complexity of this disease.
Next, the library of mutants was assessed for changes in brain activity (where the distribution of phosphorylated ERK was taken as a measure of calcium signaling) and alterations in morphology. This data has been made available in an accessible form at stackjoint.com/zbrain. While brain morphology phenotypes were only present in 12% of mutants, changes to brain activity in free swimming larvae occurred in over half of the mutants. These changes were caused by mutations in genes involved in processes previously associated with schizophrenia, such as calcium channels and synaptic pruning, but many were caused by previously ignored genes of unknown function.
One immediate use of all of this data is in the discovery of strong candidate schizophrenia genes amongst the sometimes many genes at disease-associated loci. The work also extends beyond schizophrenia to other neuropsychiatric disorders: autism and epilepsy genes were also mutated, since there is overlap between the genes associated with these diseases. Certainly, many mutants exhibited interesting phenotypes that warrant further attention.
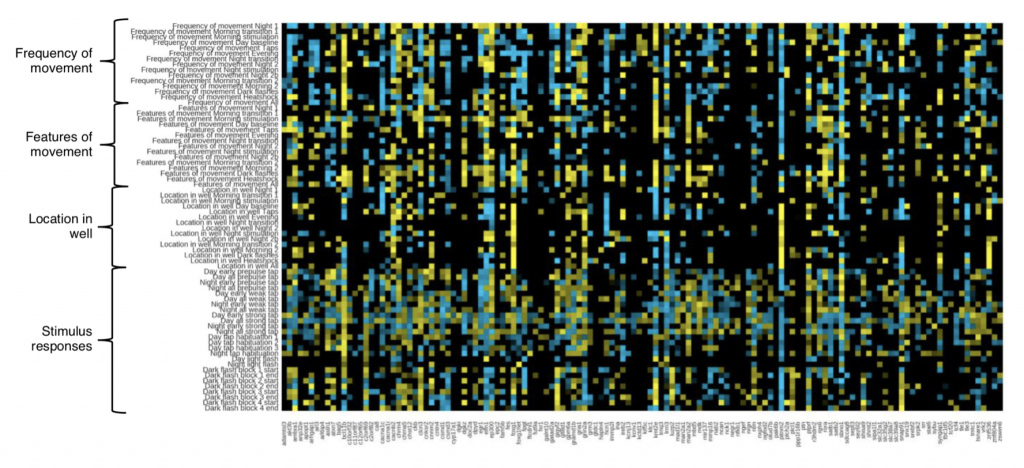
Figure 1: Summary of behavioral test results across mutant library (from Figure 2E of the preprint)
What I liked about this preprint
Our understanding of gene function in development, homeostasis and disease has mostly been advanced by manuscripts reporting on the phenotypes of one or a small number of genes at a time. One disadvantage of this approach is that phenotyping is often not standardized; different labs perform different assays or instead perform the same assays but in different ways, making comparative analysis of the effects of distinct genetic lesions complicated. Another issue is that labs naturally look at phenotypes that interest them and miss, or do not report, others. Issues like this are well understood in the mouse community, and consortia such as EUMODIC have established large-scale broad and standardized phenotyping pipelines for mouse mutants. The current preprint takes the similar approach of “many mutants/broad phenotyping”, and some of the interpretations they make were only possible because they studied so many mutants at once with standardized analysis pipelines.
This preprint also demonstrates how useful this approach is. Some unstudied genes gave unexpected phenotypes, whilst previously missed phenotypes were found in some genes that had been studied before. Happily, similarities were observed between zebrafish and mammalian phenotypes for many genes, acting as an important positive control. Given the success of this work, it is likely we’ll see others taking this approach in the study of diseases for which association data exist.
Future directions
With all this data, it is natural to ask: what next? The authors suggest how their data can be followed-up by providing proofs-of-principle for how it can generate specific hypotheses. For instance, one mutant (tcf4, a transcription factor) showed decreased activity in an area of the brain known to ‘light up’ during hunting. Upon further investigation, they indeed found reduced hunting behavior in mutants. In this way, this preprint provides a large number of entry points into detailed studies of various kinds of behavior.
Questions for the authors
For me, it would be nice to hear if the authors have any practical tips for the generation and organization of such a large mutagenesis project, accomplished in a single lab. It would also be interesting to hear their opinion on whether such large-scale approaches could in the future be used to investigate inter-(or intra-)genic regulatory sequences that may also play an important role in disease.
Further reading
Brown SDM, Holes CC, Mallon AM, Meehan TF, Smedley D and Wells S. High-throughput mouse phenomics for characterizing mammalian gene function. Nat. Rev. Genet. 2018 19(6):357-70.
McCammon JM and Sive H. Challenges in understanding psychiatric disorders and developing therapeutics: a role for zebrafish. Dis. Model Mech. 2015 8(7)647-56.
Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014 511, 421-427.
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the animal behavior and cognition category:
Cannibalism as a mechanism to offset reproductive costs in three-spined sticklebacks
Tina Nguyen
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
Trade-offs between surviving and thriving: A careful balance of physiological limitations and reproductive effort under thermal stress
Tshepiso Majelantle
Also in the genetics category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Also in the neuroscience category:
Electrophysiological correlates of conscious experiences during sleep: Lucid dreams, sleep paralysis, out-of-body experiences, and false awakenings
uMontreal Neuro preLighters et al.
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
preLists in the animal behavior and cognition category:
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Bats
A list of preprints dealing with the ecology, evolution and behavior of bats
| List by | Baheerathan Murugavel |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Also in the genetics category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (1 votes)
(1 votes)