Selectivity of Protein Interactions along the Aggregation Pathway of α-Synuclein
Posted on: 15 April 2021
Preprint posted on 29 January 2021
Article now published in Communications Biology at http://dx.doi.org/10.1038/s42003-021-02624-x
Illuminating interactomes: how Lewy Body components show previously undefined selectivity for different states of α-synuclein along the aggregation pathway
Selected by Utrecht Protein Folding and AssemblyCategories: biochemistry
Written by: Adriaan von dem Borne, Sophie Klouwer & Stijn Sijbesma
Background
α-synuclein is a protein that is abundantly expressed in the brain. Aggregation of this protein is related to the formation of Lewy Bodies (LBs), which are intracellular inclusions in which α-synuclein is the main component. It is known that certain disease-associated mutations within α-synuclein can increase its propensity to aggregate. LBs are found in the brains of patients with different types of neurodegenerative diseases such as Parkinson Disease (PD), Dementia with Lewy bodies (DLB) and in some cases of Alzheimer’s disease (AD).
LBs can contain over 70 different proteins (1). What is unknown, however, is how these interact with α-synuclein. In order to understand how LBs are formed, it is crucial to know in which order and in what form LB proteins aggregate with α-synuclein. In this study, the researchers investigate the binding of LB proteins and α-synuclein in its different forms along the aggregation pathway: monomers, oligomers and finally fibrils.
Why this preprint is interesting
We harboured a shared personal interest in LBs and PD ever since we began our investigation in the field of protein folding and assembly. Worldwide, over 10 million people are living with PD. It is through characterization of the molecular background that advances in medical sciences to treat these PD patients can ultimately be achieved. The paper describes a novel approach to understanding critical protein interactions with α-synuclein as a supramolecular complex, which may contribute to the formation of LBs. Characterizing the interactome of α-synuclein for a more holistic understanding of LB formation could fundamentally change the way in which the aforementioned neurodegenerative diseases are understood.
Key findings
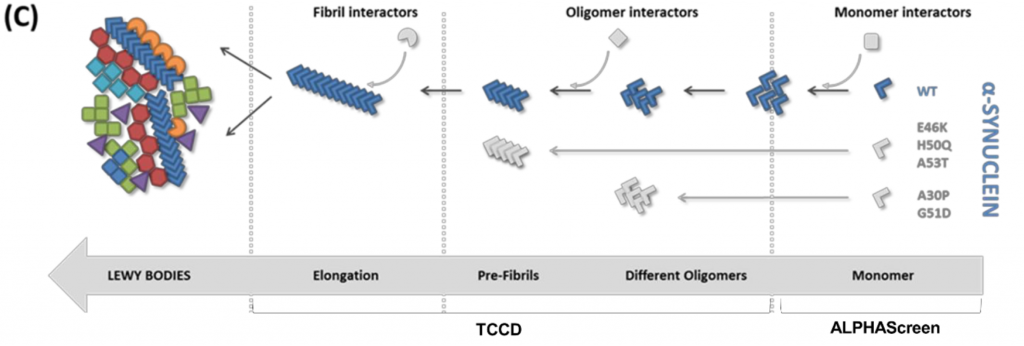
AlphaScreen on monomeric α-synuclein
The researchers start by studying the interactions of LB proteins with the monomeric form of α-synuclein. They do this using the ‘AlphaScreen’ method they previously developed (2). AlphaScreen is a highly sensitive proximity assay in which proteins are attached to donor and acceptor beads. When the interacting proteins get in close proximity, the donor transfers an excited oxygen atom to the acceptor. Because of this transfer, light will be emitted which can be detected. The researchers perform the AlphaScreen assay using mCherry-tagged α-synuclein and GFP-tagged LB proteins, which are co-expressed in a cell-free expression system. Therefore, the proteins are co-translated and can thus also interact during their folding processes. The researchers find three main interacting LB proteins from the AlphaScreen assay. These are: G-protein receptor kinase 5 (GRK5), mitogen-activated protein kinase 1 (MAPK1) and proteasome activator complex subunit 1 (PSME1). This relatively small number of hits of only three interacting proteins can be explained by the fact that other LB proteins may bind to other (non-monomeric) forms of α-synuclein.
Single molecule fluorescence: selective recognition of Lewy Body proteins by α-synuclein oligomers
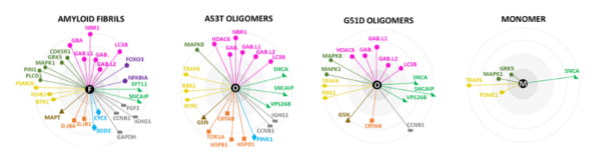
The authors then detect ‘co-assembly events’ of GFP-tagged LB proteins and mCherry-tagged α-synuclein by Two-Color Coincidence Detection (TCCD) with pathological α-synuclein mutants A30P, G51D and A53T in nanotraps. These particular mutants are chosen because of their known ability to form oligomers (3). This approach results in the observation of similar interactomes for the three mutants, and reveals that the binding to oligomeric α-synuclein occurs in sub-stoichiometric ratios. However, the small heat shock protein αB-crystallin is present in complexes of higher stoichiometry with α-synuclein. The tendency of αB-crystallin to self-aggregate may explain this result (4).
Co-aggregation or recognition of pre-formed α-synuclein oligomers?
The binding partners of α-synuclein can either bind through co-aggregation during oligomer formation, or bind through recognition of fully formed α-synuclein oligomers. In the human brain, α-synuclein oligomers will form in the presence of interacting proteins. Therefore, co-aggregation processes may play an important role. To discriminate between the two options, the authors add possible interacting proteins during α-synuclein oligomerization and after formation of these aggregate species.
The authors see that most interacting proteins do not have the ability to recognize pre-formed aggregates, meaning that recognition of α-synuclein happens during oligomerization. However, proteins from the LC3/GABARA family do recognize pre-formed α-synuclein oligomers. These proteins are involved in autophagy suggesting that this interaction may be relevant for the degradation of pathological α-synuclein.
Monitoring interacting proteins of α-synuclein fibrils
To determine the interactome of the α-synuclein fibrils, the researchers use fluorescently labelled pre-formed fibrils and add them to the possible binding partners. The authors find a high number of binding partners, including many proteins that did not co-diffuse with α-synuclein oligomers. The Tau protein is one of these interactors. In some patients suffering from neurodegenerative diseases, both α-synuclein and Tau aggregates are present. A possible connection may be that Tau monomers bind to the surface of α-synuclein fibrils, inducing nucleation of Tau aggregation (5).
The authors find that different chaperones interact with different α-synuclein aggregation states. Whereas the small heat shock protein αB-crystallin recognizes early misfolded states, Hsp40 chaperones recognize the fibrillar stages. These interactions explain why chaperones are universally present in brain inclusions like LBs.
In summary, this preprint demonstrates for the first time on a large scale that interacting proteins recognize different conformers of α-synuclein along the aggregation pathway from monomer to fibril, illuminating the pathogenic process of LB formation.
Questions
- Do you think that the point mutations could also affect binding to α-synuclein monomers as detected in the AlphaScreen assay?
- Did you observe any fibril formation after a long incubation time of the α-synuclein oligomers in the cell-free expression system? Do you think the formation of fibrils in the presence of the possible interactors may generate different results than the pre-formed fibrils did?
- Could you elaborate on why TCCD is a good method for the experiments conducted in this paper? For example how does TCCD compare to split-GFP experiments?
- Do you expect there to be common characteristics among the interacting proteins having a preference for a certain folding state of α-synuclein?
References
- Wakabayashi, K., Tanji, K., Mori, F., & Takahashi, H. (2007). The Lewy body in Parkinson’s disease: molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology : official journal of the Japanese Society of Neuropathology, 27(5), 494–506. https://doi.org/10.1111/j.1440-1789.2007.00803.x.
- Sierecki, E., Stevers, L. M., Giles, N., Polinkovsky, M. E., Moustaqil, M., Mureev, S., Johnston, W. A., Dahmer-Heath, M., Skalamera, D., Gonda, T. J., Gabrielli, B., Collins, B. M., Alexandrov, K., & Gambin, Y. (2014). Rapid Mapping of Interactions between Human SNX-BAR Proteins Measured In Vitro by AlphaScreen and Single-molecule Spectroscopy. Molecular & Cellular Proteomics, 13(9), 2233–2245. https://doi.org/10.1074/mcp.m113.037275.
- Sierecki, E., Giles, N., Bowden, Q., Polinkovsky, M.E., Steinbeck, J., Arrioti, N., Rahman, D., Bhumkar, A., Nicovich P.R., Ross, I., Parton, R.G., Böcking, T. & Gambin, Y (2016). Nanomolar oligomerization and selective co-aggregation of α-synuclein pathogenic mutants revealed by single-molecule fluorescence. Scientific Reports 6, 37630. https://doi.org/10.1038/srep37630.
- Hilton, G. R., Hochberg, G. K., Laganowsky, A., McGinnigle, S. I., Baldwin, A. J., & Benesch, J. L. (2013). C-terminal interactions mediate the quaternary dynamics of αB-crystallin. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 368(1617), 20110405. https://doi.org/10.1098/rstb.2011.0405.
- Giasson, B. I., Forman, M. S., Makoto Higuchi, M., Golbe, L.I., Graves, C. L., Kotzbauer, P. T., Trojanowski, J. Q., Lee, V. M. Y. (2003). Initiation and Synergistic Fibrillization of Tau and Alpha-Synuclein. Science, 300(5619) pp. 636-640
https://doi.org/10.1126/science.1082324.
doi: Pending
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (4 votes)
(4 votes)