Ancient origins of complex neuronal genes
Posted on: 23 October 2023
Preprint posted on 16 August 2023
Article now published in Current Biology at http://dx.doi.org/10.1016/j.cub.2024.02.021
Making sense(s) through the lens of evolution (pun intended) - old genes may drive brain evolution.
Selected by Preethi Krishnaraj, Chee Kiang EweCategories: evolutionary biology, neuroscience
Background
How does the nervous system transition from loose neuron networks in cnidarians to the complex connectomes found in vertebrates? Intriguingly, the nervous systems of many species exhibit an abundance of large genes [1]. This observation has sparked a compelling hypothesis that the expansion of gene size might be a contributing factor to the evolution of more complex brains.
During the course of evolution, the relative sizes of genes and the proteins they encode have remained relatively stable (see more below). However, in more complex organisms, a substantial proportion of the expanded genome consists of non-coding DNA. This includes introns which play vital roles in the regulation of gene expression and the generation of different protein isoforms [2].
In this preprint by McCoy and Fire, a group of genes are described that are conserved across species and potentially pivotal in driving the evolution of the nervous system. Importantly, these genes appear to have undergone significant increases in size and to have acquired a greater number of isoforms during evolution. This observation suggests that the expansion of gene size, particularly among ancient genes under high selective constraint, provides substrates for natural selection during the evolution of the nervous system.
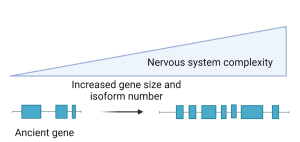
Figure 1: Gain in gene architecture complexity drives the evolution of the nervous system.
Main findings
Harmony in gene size – Relative gene size remains stable across diverse species
Many previous studies had mostly compared aggregate gene size measures across species; whereas the authors of this preprint pursued a gene-by-gene comparison approach to investigate gene size variation during evolution. Their investigation involved comparing orthologous gene sizes* across diverse eukaryotes. They explored whether gene sizes in one species correlated with those in distantly related species. Despite significant differences in absolute gene size**, they found relative gene size** to be remarkably consistent across species. This finding indicates a macroevolutionary trend in which gene sizes evolve together, regardless of absolute variations.
As an example, the authors compared orthologous genes in humans and the nematode, C. elegans. Despite consistent CDS size between each ortholog in both the species, the largest human genes were found to be 100 times larger than those in the nematode. Additionally, within both genomes, the CDS size strongly correlated with the gene size indicating a close relationship between protein and gene size on a macroevolutionary scale.
Neurological Sovereignty – Large genes often have neuronal functions
Several studies, including previous work of the preprint authors [1], have demonstrated a correlation between the substantial size of certain genes in the brain and their expression levels. In this preprint, the authors show that the top 10% largest genes include more brain-enriched genes compared to genes of other sizes, highlighting the brain’s unique gene expression patterns. Conversely, the expression of smaller genes was observed in tissues like the testis and skin.
The authors then investigated gene size distributions for specific functional categories and found that large genes often had a neuronal function, such as neuron recognition, presynaptic membrane assembly, and neuron cell-cell adhesion. These findings suggest that there are distinct types of genes: (i) those benefiting from small, condensed gene sizes including highly expressed and rapidly responsive genes, (ii) those benefiting from expanded gene sizes such as neuronal genes with multiple isoforms, and (iii) potentially a third class of genes whose sizes are influenced by unknown factors.
Timeless treasures: Most large neuronal genes are ancient
Older genes are often larger, undergo stronger purifying selection#, and evolve more slowly than newer genes. Here, the authors have detailed an analysis that focused on genes of specific ages and sizes. They found that most large genes are ancient and highly conserved, with the top 10% largest human genes averaging an age of 953 million years, while shorter genes average around 62 million years.
Further, while examining large brain-enriched genes, the authors identified their conservation not only among animals but also in basal organisms like sponges and choanoflagellates, despite lacking nervous tissues. These critical genes for nervous systems have ancient origins predating dedicated neuronal cell types.
Expanding the horizons: New isoforms evolving from large genes
In organisms characterized by expanded genomes, the authors noted the presence of ancient, highly conserved genes that were undergoing evolutionary changes by acquiring new isoforms. Specifically, this phenomenon was observed to be more pronounced in larger genes. To quantify these changes, the authors assessed the number of isoforms and compared these numbers between genes with one-to-one orthologous relationships. Interestingly, the authors noted that a group of large, ancient genes present in various species have grown even larger and more complex in vertebrates. These results indicate that large, ancient genes can actively incorporate new sequences contributing towards evolution.
What we liked about this preprint
This thought-provoking preprint proposes the interesting theory which states that increase in absolute gene size may drive the evolution of gene structures and regulatory elements, which then contribute to the diversification of the nervous systems. The bioinformatic analyses are elegant and the paper is very well written. We very much enjoyed reading it!
Questions for the authors
- We wonder if you have performed GO analysis specifically on the most ancient large genes (the 71 genes conserved between humans and sponges, for example)? Do they tend to perform a certain neuronal function?
- Your results seemingly contradict previous findings that endodermal genes tend to be older than ectodermal genes [3]. Would you be able to comment on this?
References
- Matthew J McCoy, Andrew Z Fire. Intron and gene size expansion during nervous system evolution. BMC Genomics. 2020 May 14;21(1):360. doi: 10.1186/s12864-020-6760-4.
- Warren R Francis, Gert Wörheide. Similar Ratios of Introns to Intergenic Sequence across Animal Genomes. Genome Biol Evol. 2017 Jun 1;9(6):1582-1598. doi: 10.1093/gbe/evx103.
- Tamar Hashimshony, Martin Feder, Michal Levin, Brian K. Hall, and Itai Yanai. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015 Mar 12; 519(7542): 219–222. doi: 10.1038/nature13996
Footnotes and definitions
*How did the authors define gene size? It is the length of the gene from the first to the last annotated exon, excluding untranslated regions.
**How was the gene size measured? The authors measured the size in two ways: absolute (in base pairs) and relative (ranking compared to other genes in the same genome).
What did the CDS size represent? It represented the nucleotide span within an mRNA transcript, excluding introns and untranslated regions.
How was the protein size determined? It was determined by the number of amino acids.
#purifying selection: a background selection to get rid of potentially deleterious mutations.
doi: https://doi.org/10.1242/prelights.35807
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
Also in the neuroscience category:
Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Muhammed Sinan Malik
Post-translational Tuning of Human Cortical Progenitor Neuronal Output
Jawdat Sandakly
Maturation of the glymphatic system confers innate resistance of the brain to Zika virus infection
Jimeng Li
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)