Geometric principles underlying the proliferation of a model cell system
Posted on: 7 May 2020
Preprint posted on 18 November 2019
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-020-17988-7
In a recent preprint, Wu et al. study cell-wall compromised bacteria and show that chromosome positioning and segregation is controlled by their geometry.
Selected by Martin HolubCategories: biophysics, microbiology
Background
The cell wall is evolutionary ancient and almost ubiquitous in the bacterial domain. It protects the contents of bacterial cells and largely determines their shape. About 30 genes contribute to the synthesis of cell wall material and its spatiotemporal regulation. Surprisingly, many bacteria that normally have the wall can grow and divide in its absence. The cells with compromised cell wall, L-forms, are an excellent model system for studies of the effect of cell size, shape and confinement on intracellular organization. Furthermore, the study of cells with compromised cell wall is of relevance for the development of novel antibiotics, as they often target the cell wall.
Previous work from this lab has shown that L-form growth in Gram-positive B.subtillis requires two types of mutations: i) one leading to excess membrane synthesis and ii) one that counteracts the damaging effect of reactive oxygen species that are present at elevated levels in L-forms. Interestingly, it is not clear why the reactive oxygen species occur [1,2]. Upregulation of membrane synthesis can be achieved by mutations affecting the regulation of fatty acid synthesis or indirectly by inhibiting peptidoglycan precursor synthesis. Previous studies have shown that division in L-forms is haphazard, poorly regulated, with often anucleate divisions and occasional nucleoid bisections [3]. Nevertheless, such phenotype still permits sufficient number of cells to successfully divide and results in colony growth.
The current model of division in L-forms assumes that it is driven by an imbalance between cell volume and surface area. The relative simplicity of this phenomenon makes it an attractive option for division in artificial cells and appears as a likely possibility for division of primordial cells. The systematic study of L-form divisions has been hampered by experimental issues related to time-lapse imaging of cell-wall compromised L-forms. In this paper, the authors report on the use of a novel microfluidics device for imaging elongated L-forms over extended time spans. Strikingly, they observe that the ability of cells to segregate chromosomes faithfully and symmetrically is recovered by imposing external constraints on their shape.
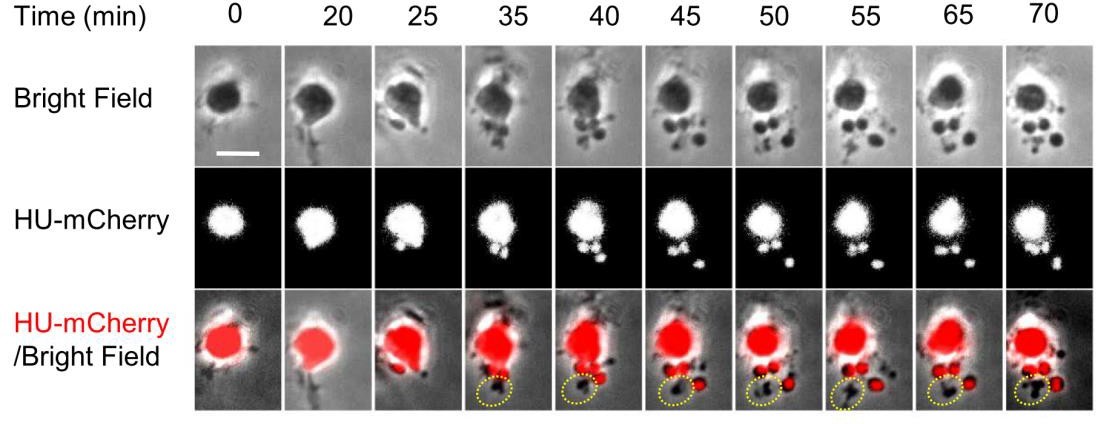
Key Findings
B.subtillis cells without a cell wall achieved robust and accurate chromosome segregation under lateral confinement that is similar to native cell width. This definitely dismisses the role of cell-wall associated phenomena in chromosome positioning. It seems that more relevant are the considerations of surface/volume, cytoplasm/nucleoid , DNA/protein and membrane/cytoplasm ratios. The authors observed a transition to regularly spaced nucleoids after both: i) initial seeding of the channel when nucleoids would overlap and ii) after passing through a region of the channel that was wider and which led back to a narrow region (diamond like shape). Despite their ability to reliably segregate chromosomes, L-forms rarely divided, even in elongated channels. The doubling time of L-forms further increased in wider channels.
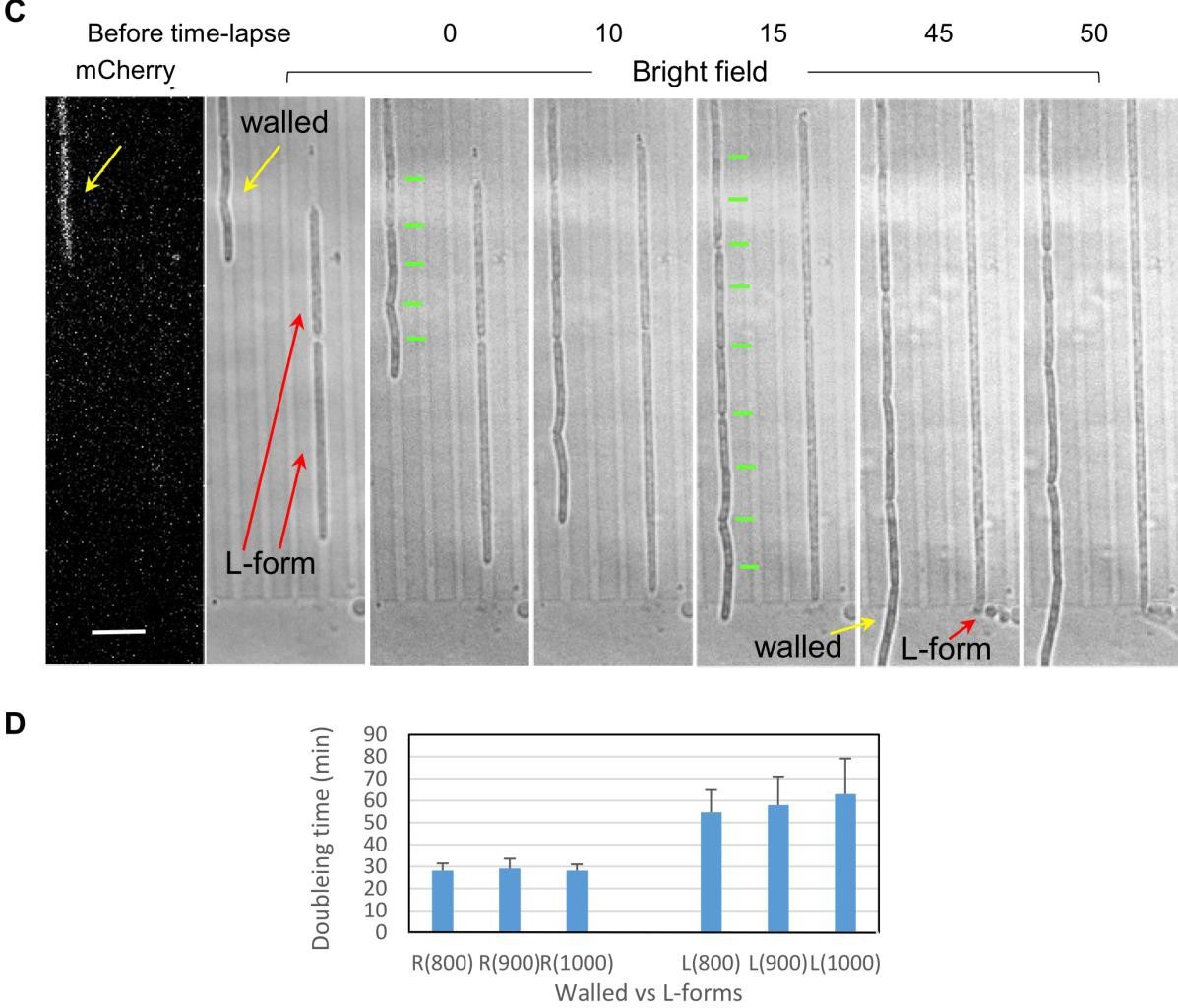
Chromosome positioning was disrupted in L-forms in the absence of tight external confinement. Chromosomes formed clumps that split only infrequently. More detailed inspection of video sequences showed that the nucleoids (or clumps thereof) show dynamic patterns of splitting and coalescence. This points to a direction of future research as the liquidity of the nucleoid suggests that it is a phase separated condensate. Nucleoid area and nucleoid centroid-to-centroid separation increased in parallel with increasing channel width, due to a failure of nucleoids to segregate and space out in wider channels. Nucleoids became wider, roughly linearly with the linear increase of channel width.
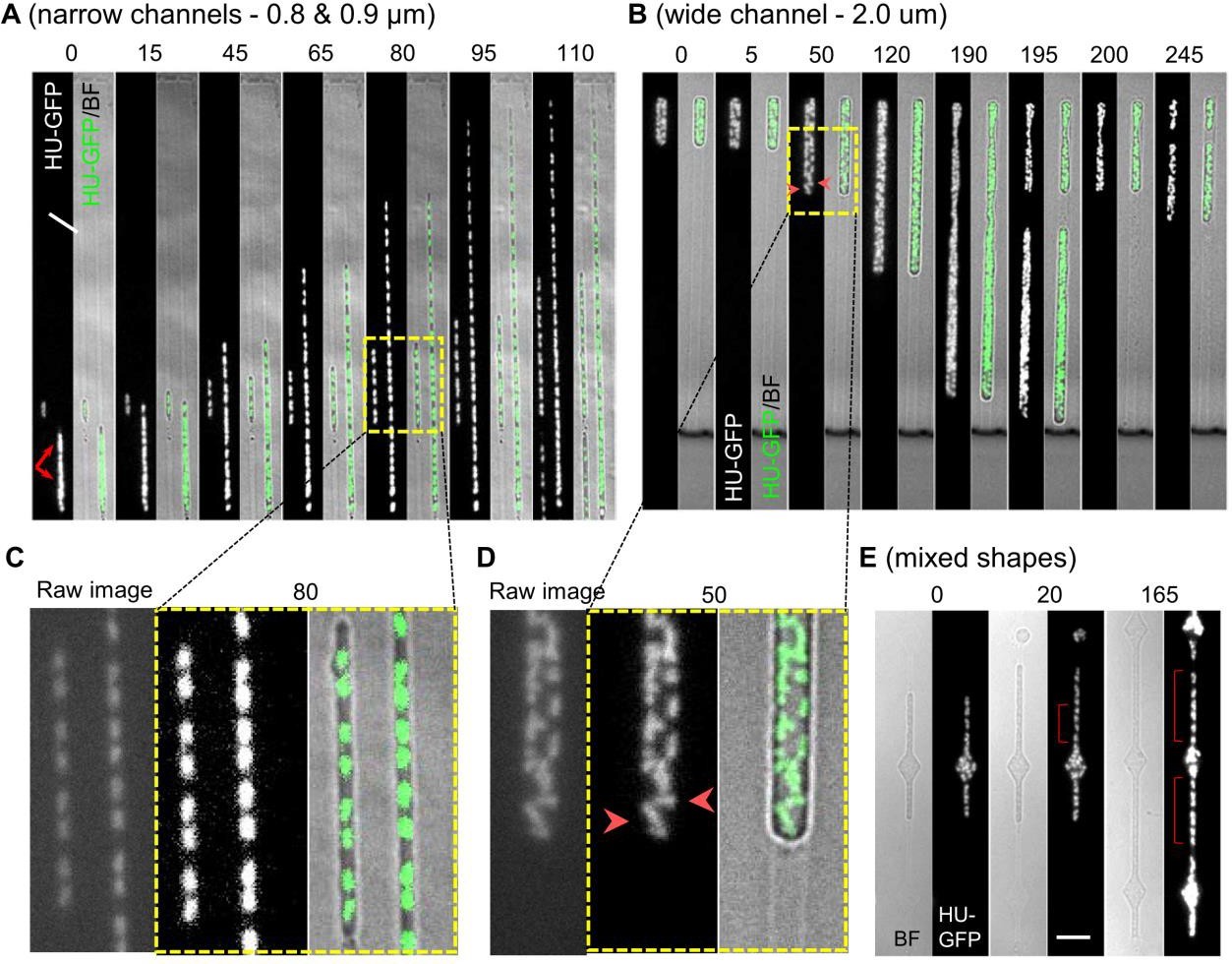
The authors observed that the division was strongly biased to inter-nucleoid spaces. This further supports “simple” biophysical and entropic arguments for chromosome segregation and positioning. Furthermore, division was successful even in cells without nucleoids. They observed the oval/sphere like objects would shed off from a cell body even at an end where there was none or little DNA. Interestingly, abolishing DNA synthesis lead to increased division frequency (in the absence of DNA) even in narrow channels that previously did not support division. This suggests that the nucleoid acts as a barrier to division. Furthermore, the authors observed that pole-localized nucleoid, resulting from asymmetric L-form division, will rapidly translocate to the midcell. They argue that such rapid response may necessitate an active repositioning mechanism.
Why I Think This Work Is Important
- Definitely excludes the role of cell-wall associated phenomena (e.g translation/transertion hypothesis) in chromosome positioning.
- Further supports the role of polymer physics and entropic argument in chromosome segregation.
- Shows that nucleoid is not necessary for budding of cells. In fact, cells bud off more easily in the absence of nucleoid, which presents and obstacle to constriction and
- Present nice microfluidic platform for studies of chromosome organization in cell-wall compromised cells.
Questions to Authors
- You comment on dynamic splitting and coalescence of nucleoid (clumps) in wider channels. Did you observe this phenomenon also in transitions from narrow channel to diamond shape chamber?
- You observe L-from division in between nucleoids. Reason could be the fact that nucleoid is probably a comparatively rigid object that needs to be bisected. Interesting question is: why does the constriction happen exactly at place where there is no nucleoid? How does the wall feel the presence of the nucleoid? Possibility would be to generate constriction seeds on some surface/volume arguments, and they would have become successful divisions only if the cut is not too costly energetically (which will be more often the case when there is a nucleoid at the cut location). What is your hypothesis on this?
- In last section of Results, you observe nucleoid repositioning from pole to mid-cell after asymmetric division. You make a suggestion of active repositioning mechanism. What is the evidence for believing there should be one, what would be plausible candidate? Can you give some arguments why entropic forces alone would not be enough to rapidly reposition the nucleoid?
- What was the speed of nucleoid movement (corrected for the growth of the bottom part)? From figure 6B, I estimate this to be around 1 µm/minute.
- In figure 3E (mixed channels), more regular spacing between nucleoids is rescued in narrow constriction after passage through wider region. From the available images, it seems that the chromosomes occur in pairs of two, but less so frequently so after the passage, and also the shape of individual chromosomes appears to be less regular. Does this observation correspond with your interpretation of the data? What is the role of the inter-diamond length in accuracy of chromosome segregation and positioning? Could it be rescued to the case of 3C, if the channel after diamond-shape was longer?
References
[1] Mercier, R.; Kawai, Y.; Errington, J. Excess Membrane Synthesis Drives a Primitive Mode of Cell Proliferation. Cell 2013, 152 (5), 997–1007. https://doi.org/10.1016/j.cell.2013.01.043.
[2] Kawai, Y.; Mercier, R.; Wu, L. J.; Domínguez-Cuevas, P.; Oshima, T.; Errington, J. Cell Growth of Wall-Free L-Form Bacteria Is Limited by Oxidative Curr Biol 2015, 25 (12), 1613–1618. https://doi.org/10.1016/j.cub.2015.04.031.
[3] Errington, L-Form Bacteria, Cell Walls and the Origins of Life. Open Biol 2013, 3 (1), 120143–120143. https://doi.org/10.1098/rsob.120143.
doi: https://doi.org/10.1242/prelights.20282
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
Also in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)