OptiJ: Open-source optical projection tomography of large organ samples
Posted on: 14 June 2019 , updated on: 24 June 2019
Preprint posted on 2 June 2019
Article now published in Scientific Reports at http://dx.doi.org/10.1038/s41598-019-52065-0
A tool for all- OptiJ: an easy-to-assemble setup and freely available software as an incentive for widescale implementation of OPT.
Selected by Mariana De NizCategories: bioengineering, biophysics
Background
Optical microscopy has been fundamental for biological discovery, however, detailed imaging within tissues is limited to a few micrometers of depth due to photon scattering (reviewed in (1)). In transparent tissues, light propagates like X rays do, so that tomographic approaches can be used for volumetric image reconstruction. In 2002, a revolutionary method was developed by Sharpe et al (2), namely optical projection tomography (OPT), to produce high resolution 3D images of fluorescent and non-fluorescent, optically cleared, biological specimens. OPT enables visualization of complete biological samples relying solely on optical sectioning. Since the publication of this method in 2002, the application of OPT within biology has expanded greatly, allowing the study of multiple processes within specimens including zebrafish, embryos, and multiple organs of rodent animals. The advent of multiple optical clearing methods has contributed greatly to the use of OPT in many organs and specimens, making it currently possible to generate and visualize volumetric data from optically transparent mice (3).
Like-wise, the generation of hybrid techniques such as OPTiSPIM, which combines selective plane illumination microscopy (SPIM) and OPT (4), has expanded the types of questions that can be addressed in multiple disciplines in biology.
While substantial technical work has been done to foster the use of these methods, a limiting factor for many labs to implement this technique is the cost and the availability of specialized software and hardware. For SPIM, this challenge was addressed by the creation of OpenSPIM in 2013 (5), a platform dedicated to making light sheet microscopy widely accessible. Furthermore, the rationale was also to allow higher throughput by making the setups more affordable, providing the possibility of using arrays to do parallel imaging. Furthermore, OpenSPIM encouraged the creation of open source solutions for SPIM image processing (6). With OptiJ, the authors address in a similar manner, the challenge of affordability and accessibility of OPT, and promote the use and further development of OPT for research by the scientific community.
Key findings and developments
- The authors have developed OptiJ, a low-cost, open-source hardware and software implementation of OPT for investigation of whole organs in 3D at near cellular resolution (Figure 1).
- This platform overcomes a key challenge: that most OPT applications require advanced technical expertise, and expensive software/hardware.
- The overall goal was to develop high-end technology that incentivises further deployment and development of OPT by the research community.
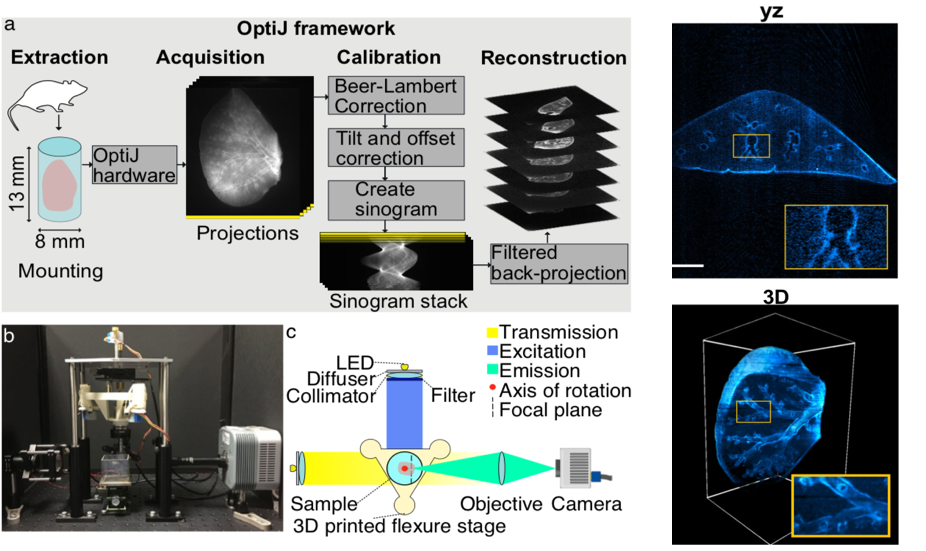
Hardware
- The OPT principle relies on the rotation of a sample to acquire 2D projections at different angles. Axis of rotation and alignment were main considerations for the setup design.
- Other main criteria guiding the component choice were: ease of access, widespread availability and low cost. With this in mind, the components developed for the setup are:
- A monolithic 3D-printed stageadapted from the Flexscope design, to accomplish the movement necessary for linear alignment and rotation of the sample with low cost motors that achieve sub-micron steps.
- A telecentric relay lens, with low numerical aperture. The telecentricity of the lens is key for an efficient filtered back projection (FBP) reconstruction approach afterwards.
- A camera. In this preprint, an Andor CLARA camera with 6.45 x 6.45 um2pixels was used, however, lower cost cameras can be used instead.
- Two broadband LEDs, emitting over a broad spectral range. A custom circuit board was designed to minimize output flicker.
- Fluorescence excitation and emission filters.
- Collimating and diffusing optics to ensure uniform illumination.
Software
- OptiJ includes a set of freely available ImageJ/Fiji plugins to process OPT data. This set of plugins is offered as an improvement over the Radon Transform (currently available in Fiji/ImageJ), as it includes calibration and accelerated reconstruction algorithms.
- Main plugins developed by the authors include:
A) Calibration plugins
- The Beer-Lambert correction plugin, which divides each transmission OPT projection (from absorption or scattering from the sample) by average brightfield image, to obtain linear attenuation coefficients corrected for non-uniform pixel intensities.
- The Estimate Tilt and Offset plugin,which tracks a fiducial marker in the projections to determine if the axis of rotation is parallel to the centre of the FOV, and produces correction values for the projection stack if not.
- The Create Sinogram plugin, which displays a Radon Transform of the projections, accounting for tilt and offset corrections. It relaxes the need for precise alignment prior to acquisitions. The output of this plugin is a sinogram.
- The dynamic offset correction plugin, which calculates a sinusoidal fit of the motion of a fiducial marker and uses the difference between the ideal fit coordinates and the actual motion in the corrected image.
B) A reconstruction plugin:
- The 2D reconstruction pluginimplements a Filtered Back-Projection algorithm to reconstruct a 3D cross-sectional stack of the original object. The plugin allows for GPU-enabled acceleration using OpenCL. This downscales the processing time from hours, to minutes.
- Implementation for the first time of the Fourier Ring Correlation (7) as a resolution measure for OPT datasets.
Proof of principle
- OptiJ allowed 3D exploration of the tertiary airways, bronchioles, and alveolar sacs in complete murine lungs, in the context of COPD.
What I like about this paper
- a) The main aim of the paper, which is to make science widely available to everyone. This is a philosophy that could permeate across many disciplines and techniques so that cost is not a limiting factor for answering research questions with the best possible technologies.
- b) That the protocols are carefully described in enough detail to allow consistent reproducibility. They consider various possible artefacts and account for them both in hardware and software.
- c) The origin of this project. It is very consistent with a philosophy of open and inclusive science, as well as fostering independence among young scientists since their early career. The work performed by the authors is the product of collaborative effort of the 14 PhD students who drove the project, as part of the programme EPSRC Centre for Doctoral Training in Sensor Technologies and Applications.
Open questions
- You have used OptiJ to image lungs as a proof of principle on the application of the setup. Previous publications have shown that various organs differ not only in optical clearance time, but also in the final clearance achievable. Do you envisage that the hardware and software setups you have implemented to calibrate the system will be equally optimal for all tissues and specimens? Under these lines, do you envisage that different mounting procedures, or optical clearance methods will influence the performance of the system?
- Given the creation of the OPTiSPIM hybrid, and the existence of OpenSPIM, is a joined low-cost platform, something you would envisage?
- Regarding the Radon Transform plugin, what are the major advances brought by OptiJ?
- You developed the Dynamic Offset correction plugin, and mention its usefulness due to the possible jitter introduced by low-cost stepper motors used for sample rotation. Can you expand further on the advantages of this method and how it works?
- You introduced the Fourier Ring Correlation in OPT. Can you expand briefly for the general readership, on how FRC has been used in various microscopy settings, as a measure of resolution without dependence on a priori calibration, and why this is novel and useful in OPT?
- For SPIM, OpenSPIM Wiki was the chosen platform to disseminate the know-how of the technique in various respects. How will you best envisage reaching your goal of a widespread access to OptiJ in the future?
References
- Ntziachristos V, Going deeper than microscopy: the optical imaging frontier in biology, Nature Methods, 2010, 7(8): 603-614.
- Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D, Optical Projection tomography as a tool for 3D microscopy and gene expression studies, Science, 2002, 296(5567): 541-545
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V, Single cell phenotyping within transparent intact tissue through whole body clearing, Cell, 2014, 158(4): 945-958
- Mayer J, Robert-Moreno A, Danuser R, Stein JV, Sharpe J, Swoger J, OPTiSPIM: integrating optical projection tomography in light sheet microscopy extends specimen characterization to non-fluorescent contrasts, Opt Lett., 2014, 39(4): 1053-1056
- Pitrone PG, Schindelin J, Stuyvenberg L, Preibisch S, Weber M, Eliceiri KW, Huisken J, Tomancak P, OpenSPIM: an open-access light-sheet microscopy platform, Nature Methods, 2013, 10(7): 598-599
- Schmied C, Stamataki E, Tomancak P, Open-source solutions for SPIMage processing, Methods Cell Biol, 2014, 123:505-529
- Banterle N, Bui KH, Lemke EA, Beck M, Fourier ring correlation as a resolution criterion for super-resolution microscopy, Struct Biol, 2013, 183(3): 363-367
doi: https://doi.org/10.1242/prelights.11202
Read preprintHave your say
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (No Ratings Yet)
(No Ratings Yet)
7 months
Kazuko dichter
Thanks for finally writing about > OptiJ: Open-source optical projection tomography of large organ samples –
preLights < Loved it!