Short N-terminal disordered regions and the proline-rich domain are major regulators of phase transitions for full-length UBQLN1, UBQLN2 and UBQLN4
Posted on: 22 October 2023 , updated on: 23 October 2023
Preprint posted on 29 September 2023
Article now published in Biophysical Journal at http://dx.doi.org/10.1016/j.bpj.2023.11.3401
Previously uncharacterized, the N-terminal region of ubiquilins is indispensable for distinct phase separation behavior.
Selected by Aniruddha DasCategories: biochemistry, biophysics
Background:
UBQLN1, UBQLN2, and UBQLN4 are three related proteins that play essential roles in the regulation of protein homeostasis. These proteins are part of a family known as ubiquilins, which are involved in various cellular processes, including the ubiquitin-proteasome system and autophagy. These processes are critical for maintaining cell health and preventing the accumulation of misfolded or damaged proteins. UBQLN1 contains a ubiquitin-like (UBL) domain and a ubiquitin-associated (UBA) domain, which allows it to bind to ubiquitinated proteins and deliver them to the proteasome for degradation. UBQLN2 is another member of the ubiquilin family, and it shares similar structural features with UBQLN1, including the UBL and UBA domains, with the exception of the proline-rich (Pxx) region. Importantly, mutations in UBQLN2 have been linked to certain cases of amyotrophic lateral sclerosis, suggesting its involvement in the pathogenesis of this neurodegenerative disorder. The third member of the ubiquilin family is UBQLN4, which also shares structural similarities with UBQLN1 and UBQLN2. This family of proteins possesses highly conserved oligomerization C-terminal STI1-II domain and putative heat-shock protein binding domain STI1-I. Here, the authors of the preprint demonstrate different phase separation behavior among ubiquilins. Also, this study shows that different epitope tags used to study the protein in vivo or in vitro could potentially influence phase separation behavior.
Importance of the study:
How do different ubiquilin proteins carry out specific functions in the cellular environment, despite their highly identical, conserved domain architecture? Here the authors describe that distinct phase separation behavior among UBQLN1, UBQLN2, and UBQLN4, could potentially allow their engagement in different physiological conditions. Moreover, the authors stress the need for careful epitope tag design and additional in vitro tests to avoid experimental artifacts while studying tagged ubiquilins in vivo.
Key findings:
UBQLN1, UBQLN2, and UBQLN4 proteins were shown to form liquid-like droplets in 200 mM NaCl. Although UBQLN1 and UBQLN2 exhibit 74% sequence identity, UBQLN2 and UBQLN4 exhibit 56% sequence identity. UBQLN2 and UBQLN4 droplets were larger than those of UBQLN1, indicating a higher phase separation propensity for UBQLN2 and UBQLN4. Also, the greater temperature dependence of UBQLN2 phase separation in comparison to UBQLN1 and UBQLN4 differentiates their condensation property.
An intrinsically disordered region (IDR) of ubiquilins – N-terminal to the UBL domain – is understudied and largely considered to be part of the UBL domain. For the first time, the authors could demonstrate that the deletion of the IDR made UBQLN1, UBQLN2, and UBQLN4 indistinguishable in terms of temperature-dependent phase separation behavior. Furthermore, the removal of the Pxx region from UBQLN2 abolished the sharp temperature dependence observed for full-length UBQLN2. Also, the addition of UBQLN2 Pxx into UBQLN4 modestly increased the temperature dependence of UBQLN4 phase separation behavior. The authors were able to show a positive regulatory role of the UBQLN2 Pxx domain and a negative role of the IDRs in guiding phase separation of ubiquilins.
The authors hypothesized that the more negative charge of UBQLN1 and the neutral charge of UBQLN2 in IDR could be a cause of the low and high phase separation propensity, respectively. They then demonstrated that the triple variant G12R/D15R/E21Q (RRQ) of UBQLN1 mimicked the UBQLN2 phase separation propensity and that the triple variant R12G/R15D/Q21E (GDE) of UBQLN2 mimicked UBQLN1 phase separation propensity.
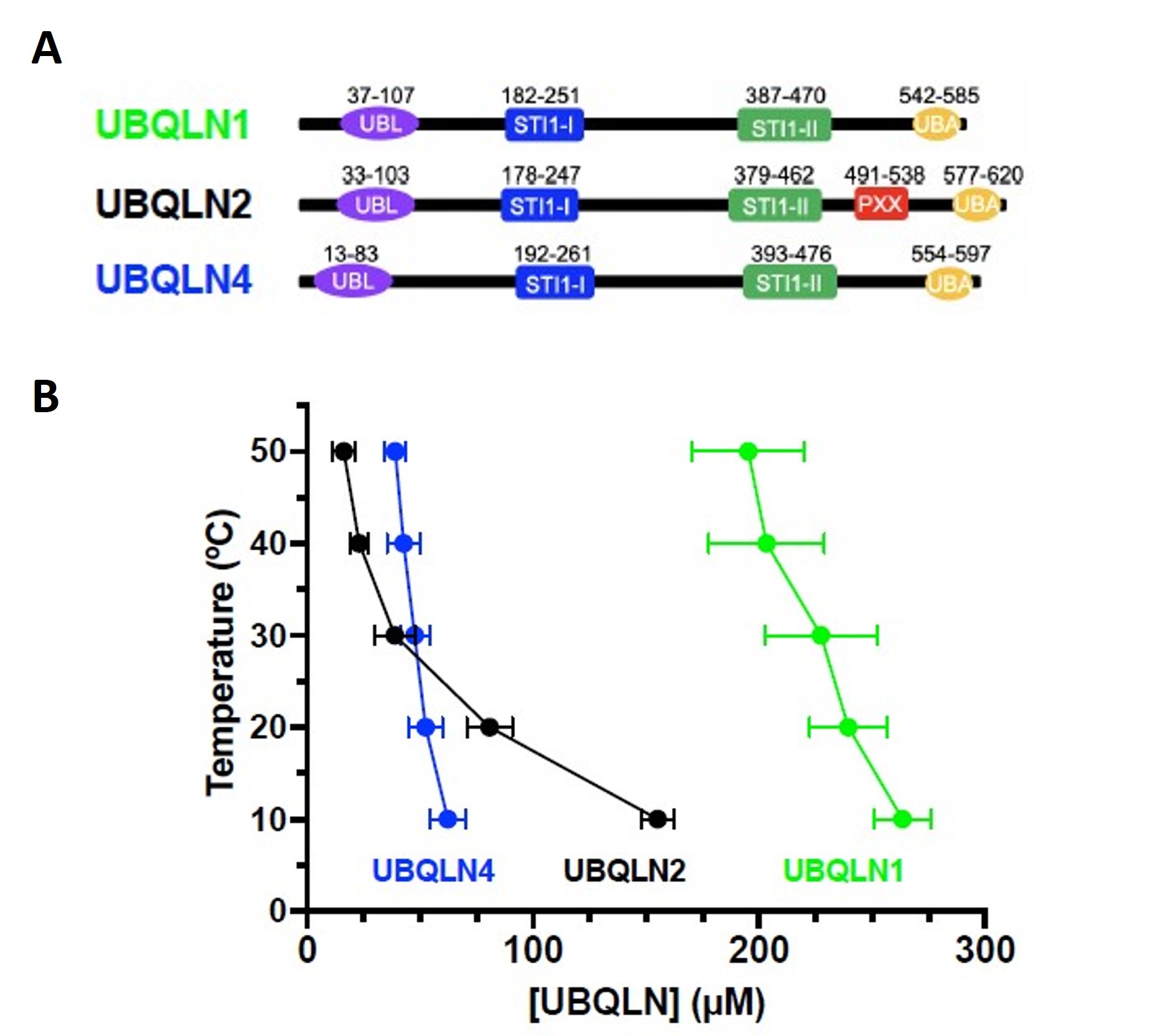
Figure 1 of Dao et al. (2023). Figure reproduced under a CC-BY-NC-ND 4.0 International License. A) Domain organization of UBQLN1, UBQLN2 and UBQLN4. B) Temperature-concentration phase diagrams for the ubiquilins by measuring saturation concentrations (csat) at different temperatures indicated that UBQLN2 and UBQLN4 more efficiently phase separate than UBQLN1.
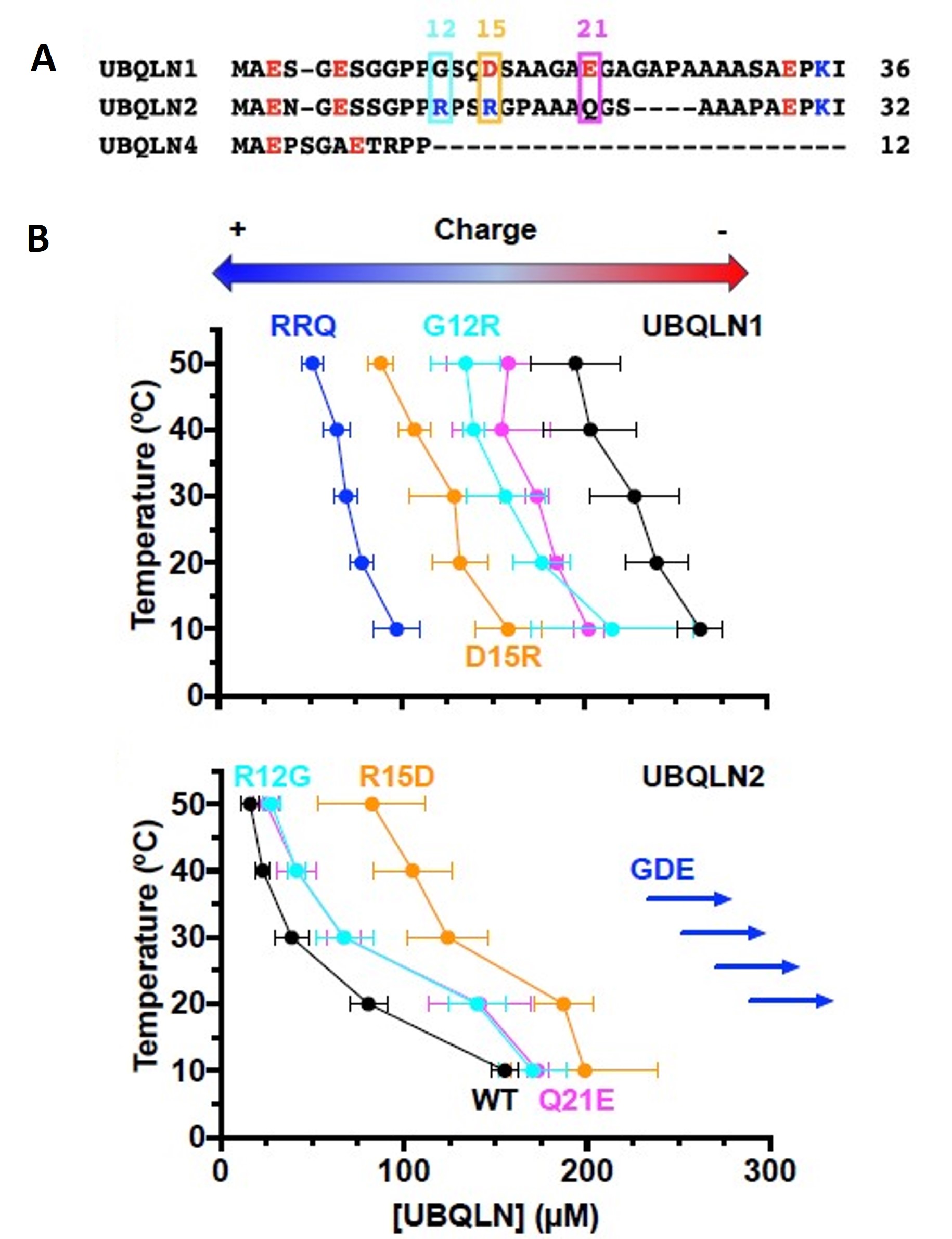
Figure 3 of Dao et al. (2023). Figure reproduced under a CC-BY-NC-ND 4.0 International License. A) Sequence alignment of N-terminal IDRs of ubiquilins showed a more negative or neutral charge for marked residues in UBQLN1, and a more positive or neutral charge for marked residues in UBQLN2. B) The phase diagram showed more negative or positive charge residue replacement in UBQLN2 or UBQLN1, respectively switching their phase separation behavior.
Often epitope tagging is critical for the in vitro or in vivo study of a given protein. Considering the sensitive phase separation behavior of ubiquilins, the authors stress that careful selection of epitope tags is very crucial. Widely used tags like FLAG and Myc could provide a higher negative charge to the protein. To avoid that, a charge-neutral ALFA tag could be potentially used to study ubiquilins. Furthermore, the presence of aromatic amino acids in the tag could improve phase separation behavior. For example, the HA tag possesses three tyrosine amino acids and has been shown to substantially increase phase separation. The authors could show that a tyrosine-free, charge-neutral tag perturbs ubiquilin phase separation the least.
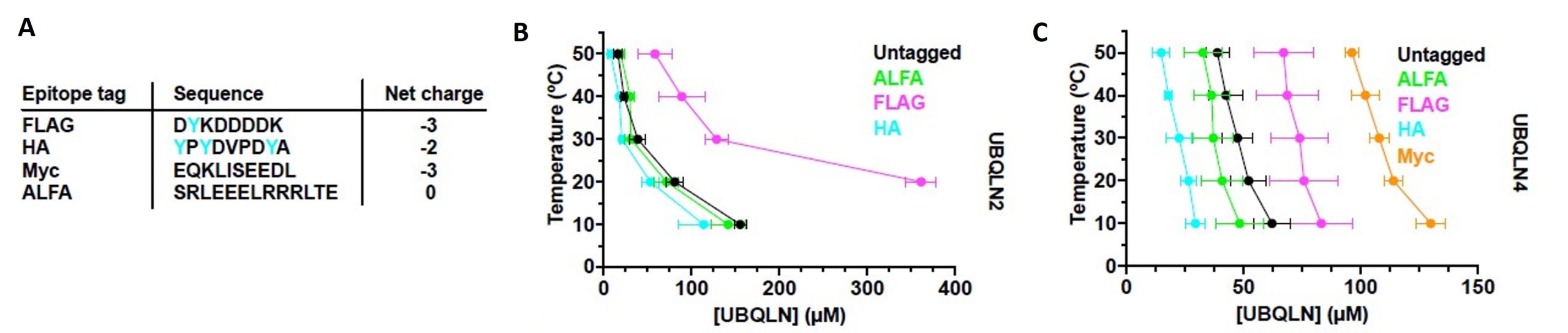
Figure 4 of Dao et al. (2023). Figure reproduced under a CC-BY-NC-ND 4.0 International License. A) Epitope tag sequences and net charges (tyrosine residues in cyan). B) Phase diagram of untagged or tagged UBQLN2, indicates that lower negative charge and higher tyrosine residues in tag positively influenced phase separation behavior. C) Phase diagram of untagged or tagged UBQLN4, indicates that lower negative charge and higher tyrosine residues in tag positively influenced phase separation behavior.
References:
Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018 Jun;28(6):420-435. doi: 10.1016/j.tcb.2018.02.004. Epub 2018 Mar 27. PMID: 29602697.
Dao TP, Kolaitis RM, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, Castañeda CA. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol Cell. 2018 Mar 15;69(6):965-978.e6. doi: 10.1016/j.molcel.2018.02.004. PMID: 29526694.
Gerson JE, Linton H, Xing J, Sutter AB, Kakos FS, Ryou J, Liggans N, Sharkey LM, Safren N, Paulson HL, Ivanova MI. Shared and divergent phase separation and aggregation properties of brain-expressed ubiquilins. Sci Rep. 2021 Jan 11;11(1):287. doi: 10.1038/s41598-020-78775-4. PMID: 33431932.
Jin X, Zhou M, Chen S, Li D, Cao X, Liu B. Effects of pH alterations on stress- and aging-induced protein phase separation. Cell Mol Life Sci. 2022 Jun 24;79(7):380. doi: 10.1007/s00018-022-04393-0. PMID: 35750966.
Riley JF, Fioramonti PJ, Rusnock AK, Hehnly H, Castañeda CA. ALS-linked mutations impair UBQLN2 stress-induced biomolecular condensate assembly in cells. J Neurochem. 2021 Oct;159(1):145-155. doi: 10.1111/jnc.15453. PMID: 34129687.
doi: https://doi.org/10.1242/prelights.35795
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Also in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (No Ratings Yet)
(No Ratings Yet)