Ultrasensitive RNA biosensors for SARS-CoV-2 detection in a simple color and luminescence assay
Posted on: 5 February 2021
Preprint posted on 8 January 2021
Colour and luminescence detection for SARS-CoV-2 that is cost-friendly and sensitive
Selected by Angika BasantCategories: molecular biology
The gold standard diagnostic method for Covid-19 infections is the RT-qPCR test, which is very sensitive, detecting viral RNA at concentrations as low as 500-1000 copies/mL (1). Nasopharyngeal samples that test positive have viral RNA at about 100,000 copies/mL on average (2). RT-qPCR relies on equipment that can be cost limiting. For example, the way the RNA is amplified for detection requires continuous cycles of temperature change for enzymes in the reaction to function and a sophisticated instrument to detect amplification in real-time. It also entails some expertise to set up and analyse results. The read-out of the technique is quantitative and would require an understanding of the reaction kinetics to be interpreted correctly.
In this preprint, the authors optimise a specialised RNA biosensor and combine it with an isothermal RNA amplification technique. Their sensor produces a colour or luminescence output in the presence of SARS-CoV-2 RNA. This approach is promising as it appears sensitive to low RNA levels and the results could be interpreted with a phone camera.
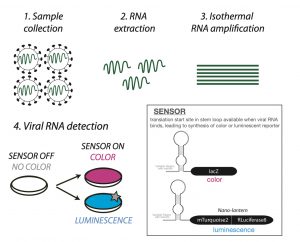
Sensing the presence of viral RNA:
Toehold RNA biosensors are long RNA molecules comprising three parts: a sequence complementary to the RNA you want to detect, a stem loop containing a ribosome binding site and a translation start site, and finally the sequence that will code for your chosen reporter protein. Such sensors have been used previously, for example in the detection of Zika virus (3). A core challenge in designing them is to choose an RNA sequence that will fold on itself to obstruct the start site in the absence of relevant RNA (sensor OFF = no reporter protein). Only when the trigger RNA is present, the sensor will bind it to allow synthesis of the reporter (sensor ON). To achieve this, the authors screened target RNA sequences in silico and in vitro to create a reliable sensor that would detect SARS-CoV-2 RNA. Their chosen biosensors recognise Orf1ab, a part of the virus genome that is invariant in >99% sequenced Covid-19 strains thus far, suggesting that this could be a widely applicable method. However, by itself this biosensor would require a high copy number (1012-1013) of RNA to work.
Amplifying RNA to make it detectable:
To amplify the RNA found in clinical samples, the authors used an isothermal amplification method that does not involve dramatic temperature cycling, called Nucleic Acid Sequence Based Amplification or NASBA (4). It involves the activity of three enzymes – a reverse transcriptase, an RNase and an RNA polymerase. Its efficiency relies on selection of optimum primers (short pieces of DNA/RNA that help initiate amplification). The authors screened several primers for NASBA to arrive at a combination that allowed their sensor to detect as low as 100 copies of starting synthetic RNA.
Testing real samples:
Next, 39 clinical samples (nasopharyngeal swabs) that had been previously tested by RT‑qPCR were analysed by this method. Rather encouragingly, the samples designated negative by RT-qPCR did not activate the sensor in this new assay, whereas the positive ones turned the sensor on i.e. the reporter gave a predicted colour shift.
A note on the read-outs:
By testing more than one reporter the authors have also demonstrated the modularity of their system. Their colorimetric reporter relies on the translation of the lacZ gene when the sensor is on. This produces an enzyme β-Galactosidase that gives a coloured product when provided an appropriate substrate. Here two substrates ONPG and CPRG were tested (5) with the latter being preferred. It gives a yellow to red colour change when the enzyme is active, and this was detectable by a cell phone camera. As an alternate, a luminescence-based reporter called nano-lantern (6) was also shown to work. This can be detected using a standard microplate reader.
What I like about this preprint:
In addition to the obvious fact that this could be an invaluable tool in the real world, I found the preprint very clear both in the experimental approach and in the way it is written. Additionally, the key computational methods used to optimise the biosensor and primers are detailed out with available code. Hopefully, this will help other researchers adopt this technique or even expand it toward other pathogens more readily.
Questions for the authors:
- What challenges do you foresee in this method being usable widely for diagnostics?
- Generally in RT-qPCR reactions, 10-1000 copies of RNA are recommended for use as template. Would you say that your technique is comparable to diagnostic RT-qPCR in terms of absolute sensitivity or are there other factors to be considered?
- This may not be practically feasible, but I was curious whether the 3 clinical samples with Ct 36-38 that tested negative in your assay could be reanalysed with one of your other sensors (1, 17, 10) and/or a different NASBA primer pair? Broadly I’m wondering if some clinical swabs may contain reaction inhibitors that impair some sensors or primers more than others, and if there is a way to address that.
References:
- Wang et al., Clinical Chemistry, Volume 66, Issue 7, 977–979 (2020)
- Wyllie et al., Engl. J. Med., 383, 1283-1286 (2020)
- Pardee et al., Cell, 165, 1255-1266 (2016)
- https://international.neb.com/applications/dna-amplification-pcr-and-qpcr/isothermal-amplification
- Serebriiskii & Golemis, Analytical Biochemistry, Volume 285, Issue 1, 1-15 (2000)
- https://blog.addgene.org/plasmids-101-imaging-with-nano-lanterns
doi: https://doi.org/10.1242/prelights.27284
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the molecular biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (1 votes)
(1 votes)