Composition and stage dynamics of mitochondrial complexes in Plasmodium falciparum
Posted on: 22 December 2020 , updated on: 23 December 2020
Preprint posted on 5 October 2020
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-021-23919-x
Categories: molecular biology
Background
Malaria parasites harbour only a single, indispensable mitochondrion with a minimalistic mitochondrial DNA encoding three proteins, COX1, COX3 and CYTB – the latter of which is the target of the antimalarial atovaquone. Although this unusual mitochondrion in Plasmodium parasites is a validated drug target, its function remains poorly understood. Due to its high sequence diversity and poor mitochondrial targeting predictions, the Plasmodium mitochondrial genome remains poorly explored. Function annotations, co-expression patterns and homology data have been integrated in silico to predict possible protein interactions, however, this approach is limited by a lack of annotated orthologues for many proteins, limited temporal resolution of expression data, and imperfect correlation between transcription and translation timing in Plasmodium species. Recent advances in methodology, including complexome profiling, has the potential of providing an inventory of protein complexes in a single experiment. It has allowed finding components of OXPHOS complexes and assembly intermediates and interactions in the mitochondria of various organisms. In their work, Evers et al (1) applied complexome profiling to map the inventory of protein complexes across the Plasmodium asexual blood and transmissible stages, enriched for mitochondria.
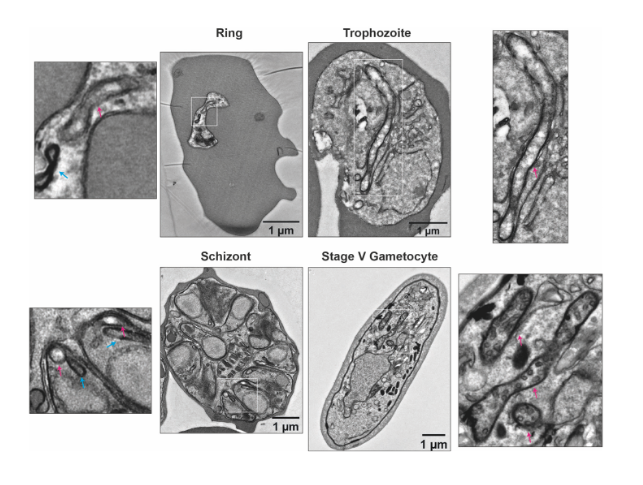
ultrastructural differences between the mitochondrion (red arrow) in asexual blood stage parasites and gametocytes. (From Ref 1.)
Key findings and developments
The authors began by demonstrating the existence of stage-specific mitochondrial ultrastructure in P. falciparum, by performing transmission electron microscopy (TEM) of the three developmental stages of asexual blood stage parasites (namely, rings, trophozoites and schizonts), and stage V gametocytes. From the obtained data, the authors point out to specific differences in cristae inside the mitochondria across the various stages. Particularly, in gametocytes, mitochondria appear more electron-dense and cover larger distinct areas, suggesting an increase in size and level of branching. The clear differences between all asexual blood stages on one side, and the gametocytes on the other, led the authors to further explore how these ultrastructural changes are reflected at the protein level. For this, the authors performed complexome profiling of mixed asexual stages and stage V gametocytes, using different enrichment methods based on syringe lysis with saponin, syringe lysis without saponin, and nitrogen cavitation, and the use of different detergents. All enrichment methods were found to be consistent, and digitonin solubilization was the chosen method for gametocyte samples. Large numbers of gametocytes were obtained by induction by conditional overexpression of GDV1 in a gametocyte producer line. Across all asexual blood stage and gametocyte samples, 1759 unique proteins were identified.
Validation of complexome profiling
In order to validate the approach, the authors verified whether other well-known, previously identified complexes could be correctly identified and whether differences existed if different isolation methods were used:
- The protein folding/degradation-involved endoplasmic reticulum membrane complex (EMC) subunits were identified (with one exception).
- The proteasome complex subunits were identified. Saponin treatment was shown to deplete all proteasome-associated assemblies, while membrane proteins were not affected.
- The rhoptry protein complex (RhopH) was identified, and a new component was described, which the authors term RhopH associated protein 1 (RhopA1).
- Six LCCL domain-containing proteins form a complex in the crystalloid (an organelle unique to Plasmodium insect stages) were identified. The authors found that in stage V gametocytes, LAP1-3 and LAP4-5 formed two distinct subcomplexes, and identified a possible LAP2-3 assembly intermediate.
- The Plasmodium translocon of exported proteins (PTEX) is a complex essential for the export of parasite proteins into the host erythrocyte. The authors identified the various components (EXP2, PTEX150, HSP101, PTEX88 and TRX2 – the latter 2 as monomers without clear co-migration with the complex being evident), as well as noting that EXP2 might be present independent from the PTEX.
The authors conclude that complexome profiling helps to distinguish the presence of proteins in different sub-assemblies, highlighting it as an advantageous tool to investigate interactions of promiscuous components or assess assembly pathways.
Investigation of mitochondrial complexes
Respiratory chain complex III (CIII)
Following the above validation, the authors next focused on the OXPHOS complexes. They found that all but one of the canonical components of cytochrome bc1 (CIII) with obvious Plasmodium orthologues, comigrated. MPPα and MPPβ, as well as 4 newly identified proteins (PF3D7_0306000, and respiratory chain complex 3 associated proteins 1-3 (C3AP1-3)) comigrated consistently with CIII subunits. The complexome profiles for CIII suggested abundant differences between asexual blood stage parasites and stage V gametocytes, with enrichment-related intensity values being 9-fold higher in gametocytes.
Respiratory chain complex IV (CIV)
So far, only 5 canonical subunits of Plasmodium cytochrome c oxidase (CIV) have been identified (COX1, COX2, COX3, COX5b and COX6b). Fragments COX2a and 2b were both identified in the complexome profiles. Recent research showed a highly divergent composition of CIV in T. gondii, containing 11 subunits specific to Apicomplexa. In their work, the authors identified orthologues for all these subunits in P. falciparum. In addition, they also identified 5 previously uncharacterized myzozan-specific proteins that consistently comigrated with the complex (which they termed respiratory chain complex 4 associated proteins 1-5 (C4AP1-5). The complexome profiles for CIV suggested abundant differences between asexual blood stage parasites and stage V gametocytes, with enrichment-related intensity values being 20-fold higher in gametocytes.
Composition of respiratory chain complexes III and IV in an evolutionary context
The authors began by mapping the gains and losses of the respective subunits along the evolutionary tree. They suggest that the three novel CIII proteins (C3AP1-3) appear to be relatively recent appearances in Apicomplexans and their close relatives. Conversely, there are proteins present in CIII from fungi and Metazoa, but absent in P. falciparum, with some being lost specifically in the Apicomplexans. Regarding the CIV complex, most of the novel proteins identified in this study appear to have myzozan origin. In addition to presence of orthologues in other species, the authors also examined addition/loss of protein domains in conserved complex membranes, and found important differences within direct evolutionary relationships.
Complex V
Classical mitochondrial function includes harnessing energy in the chemical bonds of ATP, and this process is predominantly executed by complex V (CV). The authors lysed large amounts of asexual blood stage parasites or gametocytes through nitrogen cavitation, without saponin – as this proved to be the most suitable method for this specific point. They found 14 proteins associated with CV.
Complex II
Succinate dehydrogenase couples succinate oxidation as part of the citric acid cycle to the reduction of ubiquinone in the OXPHOS pathway. CII is generally composed of at least 4 different subunits: SDHA and SDHB, catalysing succinate oxidation, and SDHC and SDHD anchoring the complex in the inner mitochondrial membrane and providing the binding pocket for haem and ubiquinone. As for CV, only the method involving nitrogen cavitation worked successfully. Although some previously suggested candidates were not found, the authors identified five putative subunits sharing a common dominant band. They assigned one of them as putative PfSDHC, and named the other 4 components, respiratory chain complex 2 associated proteins 1-4 (C2AP1-4), one of which is myzozoan-specific and plays an important role in ookinete mitochondria in P. berghei.
Protein dynamics are in line with a significant metabolic shift in P. falciparum gametocytes
Metabolomics approaches have indicated a shift in carbon metabolism in gametocytes from anaerobic glycolysis towards increased TCA cycle utilization and increased respiration. This is reflected in a general increase of mitochondrial proteins and specifically of TCA proteins in gametocytes. A further indication is the de novo appearance of cristae in gametocytes, as they typically serve as hubs for respiration. The authors investigated whether this mitochondrial phenotype would be reflected in the abundance of OXPHOS complexes. Overall, the authors found that proteins associated with CIII and CIV were significantly more abundant in gametocytes than asexual blood stage parasites. However, they also reported some variability across gametocyte samples. The authors validated their findings using complexome profiling, by using mass spectrometry, and confirmed significantly higher abundance levels of OXPHOS complex components (CII, CIII, CIV and CV) in gametocytes than in asexual blood stages. The authors also identified a few outliers (one per complex), which showed higher abundance in asexual blood stage parasites. For determining whether this trend was indicative of larger metabolic shift in gametocytes, the authors also investigated abundance dynamics of other proteins involved in central energy metabolism. They found that enzymes involved in glycolysis were more abundant in asexual blood stage parasites, while other proteins seem to be gametocyte-specific. Altogether, their data support previous metabolomic-based suggestions of a switch towards respiration and away from anaerobic glycolysis in P. falciparum gametocytes.
What I like about this preprint
I chose this preprint because it touched on a very interesting topic (i.e. mitochondrial biology) which is poorly understood in Plasmodium. As the authors emphasize in their discussion, this is surprising given that mitochondrial components are in fact antimalarial targets. I found also very interesting that they chose to compare mature gametocytes and asexual blood stages, and I like that they present an interesting method which seems to offer various advantages over others previously used. I think it will hopefully close important gaps in our knowledge of Plasmodium biology.
References
- Evers et al, Composition and stage dynamics of mitochondrial complexes in Plasmodium falciparum, bioRxiv, 2020.
doi: https://doi.org/10.1242/prelights.26595
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the molecular biology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
preLists in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (1 votes)
(1 votes)