Breaking the next Cryo-EM resolution barrier – Atomic resolution determination of proteins!
Posted on: 9 July 2020
Preprint posted on 22 May 2020
Categories: biochemistry, biophysics, synthetic biology
Background
Cryo-electron microscopy (Cryo-EM) has become very popular and successful in solving three- dimensional (3D) structures of macromolecular complexes. The technological development of electron microscopes, detectors, automated procedures as well as friendly image processing software and increasing computational power, have made cryo-EM a successful and largely expanding technology over the last decade. At resolutions better than 4 Å, atomic model building starts becoming possible but the direct visualization of true atomic positions in protein structure determination requires significantly higher resolution, which so far could not be attained by cryo-EM. In their work Yip et al (1) present a newly developed electron microscope which provides an unprecedented resolution of 1.25 Å, and for the first time allows visualization of individual atoms in a protein (apoferritin). Moreover, their setup allows also significant improvements in quality of the cryo-EM density map.
Key findings and developments
The authors explored two key questions in their work, namely whether technical improvements in hardware would allow overcoming current optical limitations in Cryo-EM, and how far such technology can be pushed.
Hardware development
The authors equipped a Thermofisher Titan Krios electron microscope (with a Falcon 3 direct electron detector), with additional electron-optical elements to increase the performance of the microscope. These elements included amonochromator and a second- generation spherical aberration corrector. The monochromator aided in reducing the energy spread of the electron beam, while the aberration corrector allowing obtaining images free of axial and off-axial coma, as well as images free of other aberrations. This setup provided significantly increased temporal coherence and less dampening of high-resolution structural details in the images. The setup also minimized linear distortions, and remains stable over longer operation times. The images obtained from this new setup, therefore, do not require post-acquisition image processing to correct these aberrations. The authors point out, however, that in data collected over several months, a small change in overall magnification occurred which needed correction. Finally, the authors discuss that in order to achieve the highest possible resolution, an extremely high quality of biological specimens is required and therefore great care must be taken in sample preparation.
Achieved resolution and image quality
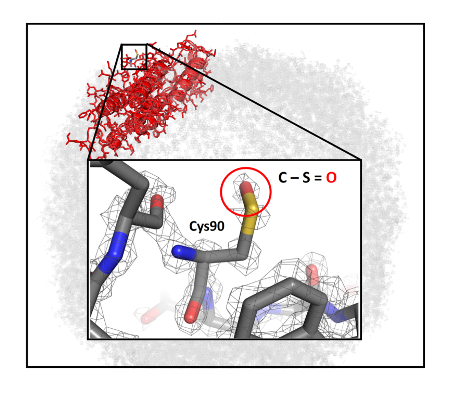
Imaged at low electron dose can result in images suffering from a significant amount of noise. To overcome this, an averaging procedure using several hundred thousand or even millions of particle images is used to calculate one 3D structure by image processing. How many such particle images are needed to obtain a specific resolution can be described by an experimental «B factor» (2). The setup hereby presented has an experimental B factor of 36 Å2. The authors predicted that using image processing alone, achieving a 1 Å resolution of apoferritin would require an unrealistic amount of time, computer power, and storage capacities. The authors went on to determine what are the particle numbers needed for the highest attainable resolution, and what features would become visible in cryo-EM maps at the highest obtained resolution with the instrument hereby presented. While an initial calculation indicated that over 5 million apoferritin particle images would be required to attain a resolution of 1.3 Å, optimization of grid preparation and imaging conditions allowed crossing the 1.5 Å resolution barrier with only 17.800 particle images. Eventually, a total number of 1.000.000 particle images allowed achieving 1.25 Å resolution. The obtained map achieved well-defined additional densities that agree with the positions of hydrogens on almost all atoms when using low thresholds for map visualization. The resolution of the map is sufficient to observe a sulfur chemical modification in apoferritin, which had not been visualized before.
In cryo-EM, the resolution of the map is estimated by the correlation of two independently calculated structures in various resolution shells in Fourier space (Fourier shell correlation (3)). The FSC provides a single number for the obtained resolution, but does not provide a direct measure of the quality of a 3D structure. Therefore, an independent means of map quality comparison is desirable. The authors compared a 1.55 Å resolution structure from a subset of the data acquired intheir instrument, with the highest resolution apoferritin structure at 1.54 Å (acquired with a different setup). They found that seven times less data was necessary with their setup, and that although the nominal resolution was similar, a significant improvement in map quality was achieved with the setup hereby presented.
The authors then discuss improvements beyond their setup, which would be desirable in new EM hardware. Improvements discussed include a) improvement in image recording speed by faster cameras and optimized data acquisition schemes; b) next generation detectors which could potentially improve the “B factor” to <30 Å2 , needed to break the 1 Å resolution barrier with a manageable amount of data. Altogether, the authors conclude that improving electron microscope hardware is essential for Cryo-EM in terms of resolution and throughput.
What I like about this preprint
I chose this preprint because the achievement is completely novel. I think technology improvements have led the way and preceded many important and paradigm-shifting biological discoveries. I liked the questions the authors explored and the ones raised for the future. I enjoyed reading the preprint, and I think this achievement will have great impact in various disciplines.
Open questions
- I enjoyed a lot reading your preprint. I was wondering if you could expand further (for experts and non-experts) on the applications for which your improved setup could be applied? It’s opening a whole new set of questions..
- You mention in your preprint that despite the improvements your setup achieves, further improvement can be reached to achieve high throughput and higher resolution. Initially one of the questions you raised was whether improvements can come solely from the hardware side. Software developments seem to be taking place also at a high pace. Do you envisage a combination of hardware and software will allow the further achievements you discuss? If so, what are main factors to improve when it comes to software that you can jointly address with the hardware improvements you are exploring?
- Regarding the further hardware improvements you discuss, can these be implemented in the setup you present here?
References
- Yip KM, et al Breaking the next Cryo-EM resolution barrier- atomic resolution determination of proteins, bioRxiv, 2020.
- Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of Molecular Biology 333, 721-745, 2003.
- Harauz, G. & van Heel, M. Exact filters for general geometry three dimensional reconstruction. Optik (Stuttgart) 73, 146-156, 1986.
doi: https://doi.org/10.1242/prelights.22687
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Also in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
Also in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)