Engineered Enzymes that Retain and Regenerate their Cofactors Enable Continuous-Flow Biocatalysis
Posted on: 22 March 2019 , updated on: 29 September 2019
Preprint posted on 5 March 2019
Article now published in Nature Catalysis at http://dx.doi.org/10.1038/s41929-019-0353-0
How far it’ll flow, how far it’ll go: the use of engineered enzymes in continuous-flow catalytic biosynthesis
Selected by Zhang-He GohCategories: bioengineering, synthetic biology
Background of preprint
Despite researchers’ ability to immobilise enzymes for continuous-flow applications, most continuous-flow applications rely heavily on cofactor-independent enzymes. The difficulty in using cofactor-dependent enzymes is twofold: the cofactor needs to be recycled for synthetic feasibility on an industrial scale; yet the recycling of the cofactor needs it to be able to diffuse throughout the reactor. This precludes the use of cofactors (and thus cofactor-dependent enzymes) in continuous-flow reactors due to the current lack of an engineering workaround. Therefore, in this preprint, Hartley et al. provide a proof-of-concept for the incorporation of cofactor-dependent enzymes into biocatalytic continuous-flow reactors.
Key findings of preprint
The findings of this preprint by Hartley et al. can be divided into three parts: (A) design of the “nanomachine”, which refers to the authors’ biocatalyst that retains and recycles its cofactor; (B) assembly of nanomachine; and (C) characterisation of the nanomachine’s function.
(A) Design of nanomachine
Inspired by enzymes that retain their substrates via covalent attachment during the reaction cascade, Hartley et al. invented a nanomachine with three domains (Figure 1). The first, a cofactor-dependent catalyst, carries out the desired chemical reaction. The second domain helps to recycle that cofactor catalyst. The last domain is an esterase conjugation domain that allows the immobilisation of the whole nanomachine to a surface. These three domains were encoded by a single gene for production in E. coli with a short spacer of 2-20 amino acids between each domain. The cofactor was modified and conjugated to the spacer between the catalytic and cofactor recycling modules using a flexible maleimide-functionalised polyethylene glycol (PEG) linker. This PEG linker allowed the modified cofactor to move between sites, thus overcoming the problem of cofactor diffusion faced by incorporating cofactor-dependent enzymes in continuous-flow bioreactors.
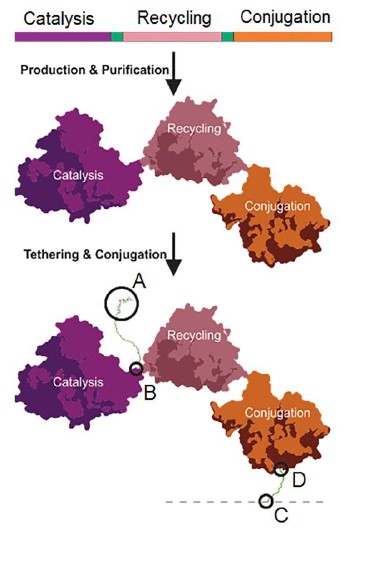
Figure 1. The structure of the nanofactory in the preprint by Hartley et al.. Reproduced from the original preprint by Hartley et al. under a CC BY-NC-ND 4.0 licence.
(B) Assembly of nanomachine
Due to the challenge in synthesising the highly chiral D-fagomine using non-biological catalysts, Hartley et al. decided to work on this synthesis for their proof-of-concept in this preprint. For the key steps in the assembly of the nanomachine, Hartley et al. chose the respective enzymes for their compatibility in batch reactions, high catalytic rates, relatively simple quaternary structures, and thermostability (Table 1). For the conjugation module, the authors chose a serine hydrolase enzyme coupled with the suicide inhibitor trifluoroketone to catalyse the formation of a site-specific and stable covalent bond between the inhibitor and the catalytic serine residue.
Table 1. Key steps in D-fagomine synthesis, their accompanying enzymes, and their respective yields. Parts of the nanofactory are highlighted in orange.
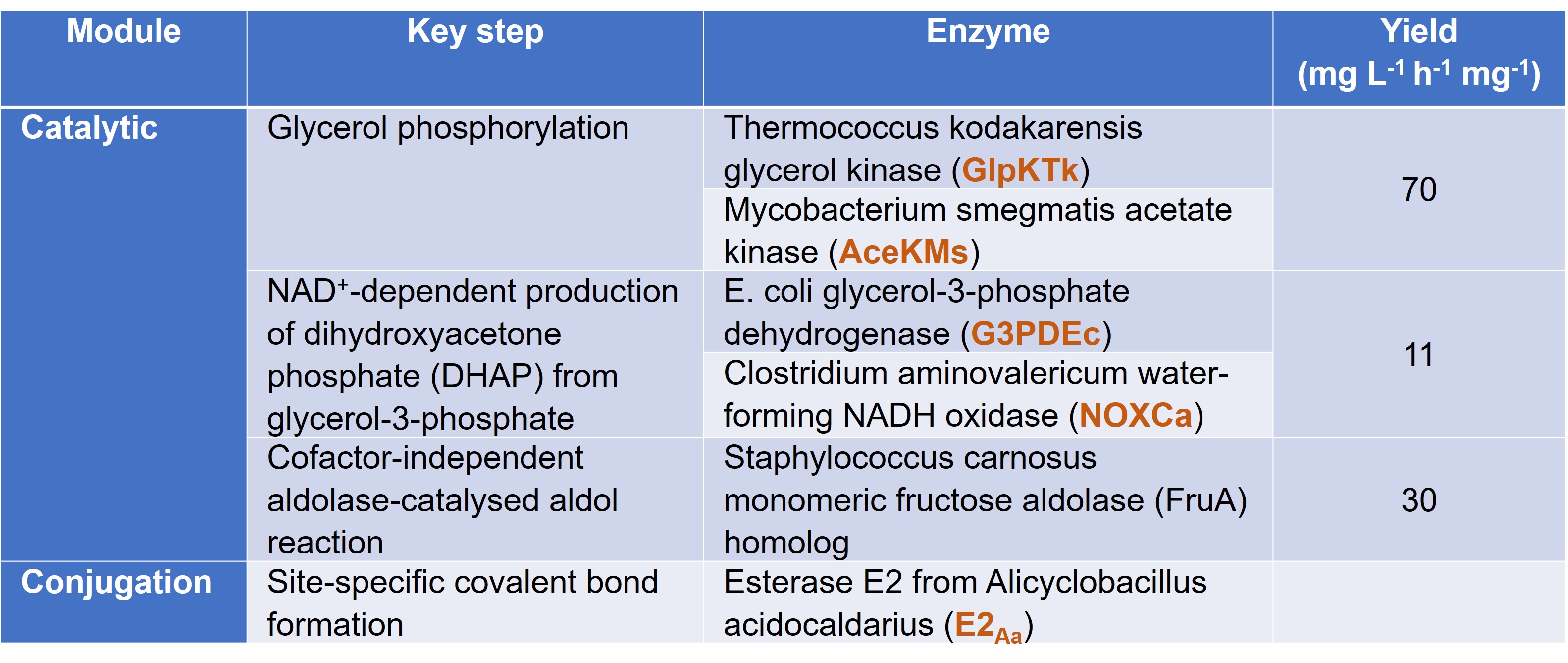
In synthesising the nanofactory, the authors created two gene fusions: the first contained the genes for GlpKTk (N-terminus) and for AceKMs (C-terminus); while the second contained the genes for G3PDEc (N-terminus) and NOXCa (C-terminus). For each of these fusions, the authors fused E2Aa to the respective C-termini. Importantly, the authors verified that their fused proteins largely retained their original catalytic functions as measured by the affinity for the enzyme for its substrate (kM) and the catalytic efficiency of the enzyme (kcat). This led Hartley et al. to conclude that their technique may be applicable to other biocatalytic systems as well.
(C) Characterisation of nanofactory
In the third section of their preprint, Hartley et al. found that their nanomachine reactors were able to synthesise products in high yields (Table 1). This observation could be ascribed to their use of a continuous flow reaction, which allowed the authors to mitigate the two limiting factors in these reactions: (i) product inhibition and (ii) equilibrium control.
Hartley et al. also characterised the turnover number (also known as the catalytic efficiency of the enzyme, kcat) for the respective cofactors. Because the turnover number of the cofactors in both reactions was identified to be limited by the inactivation of the domain rather than losses of the cofactors themselves, Hartley et al. posit that this could be improved by enhancing the stability of the enzymes involved.
What I like about this preprint
I selected this preprint for two reasons. The first is the significance behind the work: Hartley et al. present a solution to the challenge of using cofactor-dependent enzymes in continuous flow synthesis. Specifically, the authors describe the invention of a nanofactory that consists of three linked modules, along with a conjugated modified cofactor that can be recycled throughout the synthesis.
Second, the work is also elegant in its own way—to paraphrase Moana, everything is by design, and every component in the nanofactory has a role in that nanofactory. The authors have documented their painstaking efforts in the supplementary section, which is certainly worth a read: just to name a few, these experiments include (i) transformation of the cells, (ii) synthesis and purification of the enzymes, and (iii) characterisation of enzymatic activity using various assays.
Future directions
The modular aspect of the system will probably lend itself to other applications, and future directions are likely to develop in the following three ways:
- Application of the nanofactory’s modular approach to other synthetic challenges. The same principle behind the nanofactory may be adapted to link different combinations and permutations of other enzymes together. This would enable scientists and engineers to perform series of other stereoselective syntheses. While it is conceivable that this method may be applied to achieve a near-infinitesimal number of synthetic routes, such an ambition may be tempered by biological limitations. For example, as transformation efficiencies of coli decrease as the size of the plasmid to be inserted increases, there may be a practical maximum to the gene size which can be transformed into the E. coli.1 As a result, there may be an upper limit to the number of modules that can be incorporated into a single nanofactory produced by E. coli. Therefore, larger nanofactories may need be achieved through other biotechnological strategies and by using other producing organisms, such as Chinese Hamster Ovary (CHO) mammalian cells.
- Optimisation of the nanofactory’s modular approach to boost yields. Hartley et al. point out that stability is likely the main factor that limits the catalytic efficiency, kcat, of the enzymes. Therefore, future work will involve the enhancement of the stability of these enzymes.
- Optimisation of the individual enzymatic modules in the nanofactory. Due to the multifactorial nature of the conditions in the nanofactory, there is plenty of room for optimisation beyond the extensive work already undertaken by the preprint authors. Factors such as the peptide spacer of 2-20 amino acids between the modules, temperature, and flow rate are all examples of aspects that could be further optimised to boost yields of this nanofactory. This is especially the case for advances in biotechnology, in which a balance must be struck between considerations beyond the optimisation of the desired protein itself—these may include the growth rate of host cells that express the desired protein, as well as the expression levels of the desired protein in host cells.
All in all, the potential applications of such a technology are especially exciting. Watch how far it flows.
References
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology 1983, 166, 557-580.
doi: https://doi.org/10.1242/prelights.9579
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)