Guide RNA categorization enables target site choice in Tn7-CRISPR-Cas transposons
Posted on: 16 July 2020
Preprint posted on 2 July 2020
Article now published in Cell at http://dx.doi.org/10.1016/j.cell.2020.11.005
Tn7 transposon with a private guide: CRISPR-Cas type I-F3b utilizes a specialized guide RNA for site-specific transposition
Selected by haridha shivramCategories: bioinformatics, evolutionary biology, genetics, genomics, microbiology
Background:
CRISPR-Cas is a prokaryotic defense system that provides its host with adaptive immunity against invading mobile genetic elements. In addition to protein-coding genes that function at different stages of the CRISPR-Cas pathway, the CRISPR locus contains an array of repeats flanked by variable sequences called spacers. These spacers serve as an immunological memory of past infections by encoding for a sequence corresponding to a genomic segment of the infecting element. During interference, the CRISPR effector complex is guided to cleave the target nucleic acid by a non-coding RNA (guide RNA) formed by a pair of repeat and spacer.
Several decades of research on these systems have uncovered multiple types of CRISPR-Cas systems (Type I-VI) that vary in their composition and mechanism [1]. In 2017, Peters et al bioinformatically identified ~400 instances that includes a single ancestral association between the type I-F CRISPR-Cas system and a bacterial transposon, related to a specialized element called Tn7 [2]. In addition to association with type I-F, now categorized as type I-F3, two evolutionarily independent associations of Tn7 with type I-B and V-K CRISPR-Cas systems have been identified [2,3]. These Tn7 associated CRISPR-Cas systems are unique in that they lack the integrases (Cas1 and Cas2) that are involved in acquisition of new spacers. Additionally, type I-F3 also lacks the Cas3 endonuclease activity that is responsible for target DNA degradation [2].
Tn7 is a cut-and-paste DNA transposon that can undergo transposition into random integration sites in a sequence-independent manner, while specifically targeting mobile plasmids (TnsABC +E dependent pathway) or can site-specifically integrate at sites proximal to conserved protein coding genes (TniQ/TnsD dependent pathway). Typically, integration of Tn7 elements into unique sites in the TniQ/TnsD-dependent pathway is thought to be driven by the evolution of the targeting specificity of TnsD/TniQ DNA binding proteins. However, with the discovery of CRISPR-associated Tn7 systems, it was speculated that the site-specific integration of these systems might be achieved through the guide RNA encoded within the CRISPR-Cas system. Last year, Klompe et al tested this hypothesis by reconstituting the Vibrio cholerae encoded Tn6677 system in Escherichia coli and demonstrated that the transposon system can be targeted to unique sites based on the guide RNA sequence [4]. Although this suggested that the Tn7-like transposon has repurposed the CRISPR-Cas system to integrate site-specifically in a guide RNA dependent manner, no target sequences corresponding to the I-F3 spacers were previously identified around the integration sites. In this preprint, Petassi et al not only provide evidence for guide RNA driven integration of type I-F3 CRISPR system, but also reveal several interesting features of these systems.
Major findings:
Guide RNA dependent integration of type I-F3 locus
Through data mining of ~53000 genomes, the authors identified ~801 Tn7-like elements containing the type I-F3 CRISPR-Cas variant that can further be classified into type I-F3a and IF3b. Strikingly, for every integration event the authors were able to identify the complementary target sequences corresponding to one of the spacers from the array. Several interesting observations were made about these interactions: (1) the spacer interaction almost always occurred ~48bp away from the right end of the transposon. (2) the transposition-associated spacer (taSpacer) always appeared to be the last spacer in the array. In type I-F3a the taSpacer with its flanking repeats occurred after a 70-90 bp gap in the array, while for I-F3b, the array appeared to be contiguous (Fig. 1A). (3) In contrast to typical repeats that are nearly identical to each other, the repeat flanking the taSpacer (atypical repeat) showed significant sequence divergence compared to the other repeats in the array.
Atypical repeats form guide RNA complexes with enhanced transposition capabilities
To determine if the atypical repeats were capable of forming functional guide RNA complexes and drive transposition, the authors performed transposition assays where they compared transposition frequencies into the genome or into exogenously introduced plasmids for different spacer sequences flanked by either typical or atypical repeats. Surprisingly for I-F3b, they found that not only were the atypical repeats capable of forming functional guide RNA complexes but the I-F3b elements also showed higher transposition frequencies when the taSpacers were flanked by atypical repeats compared to typical repeats. The transposition frequencies however drastically varied based on the spacer sequence making it difficult to predict the outcome of guide RNA dependent transposition events. In contrast to I-F3b, atypical repeats did not lead to higher transposition frequencies for type I-F3a system. What could lead to such functional differences between these two classes of Tn7 CRISPR systems? To answer this, future work detailing the differences in the mechanism of guide RNA dependent transposition will be critical. It would be interesting to see how the divergence observed for atypical repeats impact the Cas effector conformation and its interaction dynamics.
Type I-F3b CRISPR-Cas system utilizes specialized guide RNAs for transposition
Since the typical repeats of type I-F3 and canonical type I-F are highly similar, the authors next tested if the type I-F system could use the I-F3 arrays for interference. If that were possible, the type I-F CRISPR complexes could be guided by the chromosome targeting taSpacers to self-target the host’s own genome for degradation leading to autoimmunity. To test this, the authors performed plasmid transformation assays with the canonical type I-F system using typical and atypical repeats from type I-F3 arrays. Based on this assay, typical repeats but not the atypical repeats from type I-Fb arrays were capable of guiding interference by the canonical type I-F system. This again was not the case for type I-F3a suggesting that if left unchecked, the cohabitation of type I-F3a and canonical type I-F systems could lead to autoimmunity (Fig. 1B).
How might type I-F3a be preventing autoimmunity? Spacers targeting the host’s own chromosome is not unique to the Tn7 CRISPR systems. Several genomes have been identified that encode such self-targeting spacers [4]. In fact, the self-targeting property of some of these genomes have been used as clues to identify mechanisms to control CRISPR activity. Many Anti-CRISPR proteins that directly counteract CRISPR-Cas interference machinery have been discovered in these self-targeting genomes [5]. Although the Authors did not find any of the known Anti-CRISPRs to be conserved and widespread in the type I-F3 genomes, these genomes might encode novel Anti-CRISPRs.
In addition to guide RNAs being specialized for transposition at least for type I-F3b, both classes of Tn7 CRISPR-Cas systems are transcriptionally controlled by the transcription factor, XRE. For type I-F3a, the XRE directly interacts with the promoter upstream of the array containing the spacer involved in transposition. While for type I-F3b, XRE interacts with the promoter upstream of Cas genes (Fig. 1C). It is possible that for type I-F3a, the XRE represses expression of the array required for transposition to mitigate autoimmunity. Alternatively, consistent with the observation that XRE represses expression of the Cas operon in type I-F3b, it also may be that the Tn7 CRISPR-Cas systems are under self-regulation to limit the time window of transposition.
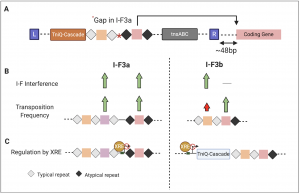
Fig 1. Summary of some of the key findings highlighting the differences between type I-F3a and b Tn7 CRISPR-Cas systems. (A) Schematic showing the locus structure of Tn7-CRISPR-Cas system depicting its integration proximal to a conserved gene with the spacer interaction site highlighted by the arrow. (B) For type I-F3a, the spacers flanked by typical or atypical repeats are equally efficient for transposition and are also capable of being used for interference by canonical type I-F CRISPR-Cas system. In case of type I-F3b, spacers flanked by atypical repeats are specialized for guiding transposition and cannot be utilized by type I-F CRISPR-Cas system for interference. (C) XRE represses transcription of both type I-F3a and I-F3b locus but vary in the site of interaction.
Follow-up questions
- The presence of guide RNA targeting sequence strictly found ~48bp proximal to the integration site in addition to its requirement for targeting the integration site is a highly compelling evidence for guide RNA driven transposition. It is intriguing however that the guide RNA directed transposition could tolerate ~10 mismatches.
- Is this tolerance for mismatches also spacer sequence specific? Would increasing the number of mismatches abolish transposition? What is the threshold?
- How many other sequences complementary to the transposition-associated spacer (taSpacer) can be identified in the genome (Allowing 10 mismatches)? If multiple possible interaction sites exist, could it mean additional factors might be involved in integration site specificity?
- It is interesting that the spacer used for transposition is always encoded at the end of the array. This could either be because the spacer is functional for transposition only when organized at the end of the array. Alternatively, it could mean that the type I-F3 system was introduced into the host through transposition first, and then with time new spacers got acquired. Is it critical that the spacer be present at the end of the array to be functional in guiding transposition? In the experimental setup shown in Fig. 3A-B, would switching the R2 repeat to an atypical repeat R3 lead to a rescue in transposition frequency of S1 into pS44-1 plasmid?
References:
- Makarova, K.S., Wolf, Y.I., Iranzo, J. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83 (2020). https://doi.org/10.1038/s41579-019-0299-x
- E. Peters, K.S. Makarova, S. Shmakov, E.V. Koonin Recruitment of CRISPR-Cas systems by Tn7-like transposons Proc. Natl. Acad. Sci. USA, 114 (2017), pp. E7358-E7366
- Strecker, J., Ladha, A., Gardner, Z., Schmid-Burgk, J.L., Makarova, K.S., Koonin, E.V., and Zhang, F. RNA-guided DNA insertion with CRISPR-associated transposases. Science 1054 365, 48-53 (2019).
- Klompe, S.E., Vo, P.L.H., Halpin-Healy, T.S. et al. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). https://doi.org/10.1038/s41586-019-1323-z
- Watters KE, Fellmann C, Bai HB, Ren SM & Doudna JA Systematic discovery of natural CRISPRCas12a inhibitors. Science (80-. ) eaau5138 (2018). doi:10.1126/science.aau5138
doi: https://doi.org/10.1242/prelights.23161
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Computational design of pH-sensitive binders
Mohammed JALLOH
Longitudinal single cell RNA-sequencing reveals evolution of micro- and macro-states in chronic myeloid leukemia
Charis Qi
Also in the evolutionary biology category:
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
Beyond venomous fangs: Uloboridae spiders have lost their venom but not their toxicity
Daniel Fernando Reyes Enríquez, Marcus Oliveira
Also in the genetics category:
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
VANGL2 shapes the mouse heart tube from adjacent epithelia and without planar polarity
Anubhav Prakash
Also in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the evolutionary biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the genetics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Also in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)