Lipid bilayer thinning near a ubiquitin ligase selects ER membrane proteins for degradation
Posted on: 21 November 2025 , updated on: 25 November 2025
Preprint posted on 1 November 2025
Exposing hydrophilic residues pushes misfolded ER proteins towards degradation.
Selected by Ana Sanchez-MolinaCategories: molecular biology
Why this new work is important
This study provides a clear model for how cells monitor the quality of their membrane proteins. The work reveals a simple, physics-driven mechanism: rather than being recognized by a specific molecular sensor, faulty proteins are identified through basic energetics and membrane dynamics. It also uncovers a key difference between single-pass and multi-pass membrane proteins.
By introducing the idea that local membrane thinning acts as a functional gateway for protein movement, this study reshapes our understanding of protein quality control and may have broader implications for other membrane systems, such as mitochondria or Golgi.
Background
Many proteins end up in the endoplasmic reticulum (ER), where they are either inserted into the membrane or folded inside the lumen with the help of the translocon. Once there, they face strict quality control: only properly folded and assembled proteins can stay. If a protein fails to fold correctly, it is sent for degradation through the ER-associated degradation (ERAD) pathway, a conserved process across all eukaryotes that prevents toxic buildup of misfolded proteins. During ERAD, defective proteins are tagged with ubiquitin, extracted into the cytosol, and degraded by the proteasome.
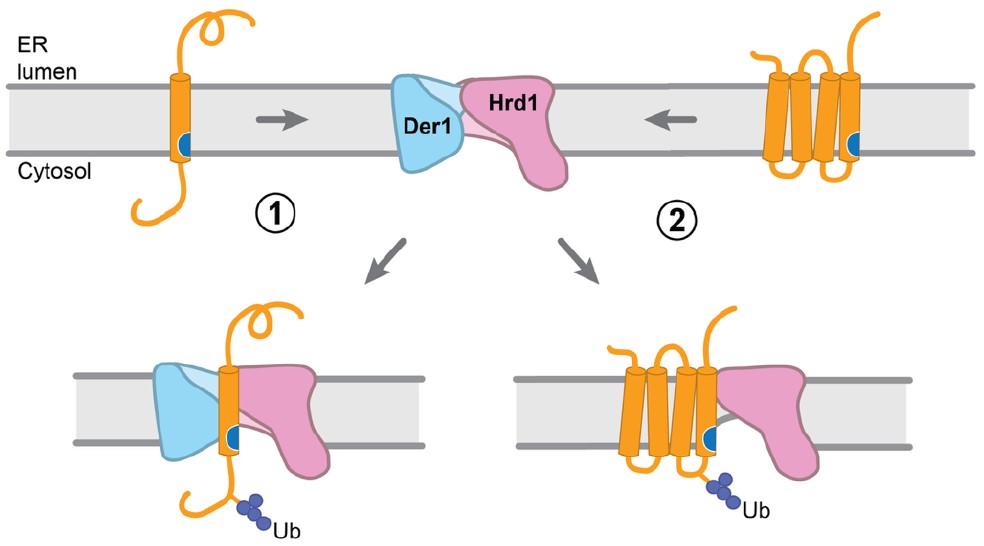
For misfolded luminal proteins, the process is relatively well understood. In yeast, it involves the Hrd1 complex, where Hrd1, a ubiquitin ligase, and its partner Der1 help move misfolded polypeptides through a locally thinned region of the ER membrane. However, the mechanism for membrane protein degradation has been less clear, and this study sheds light on that process. The authors show, using budding yeast, that misfolded membrane proteins are selected for degradation through a general mechanism in which they partition into a locally thinned region of the ER membrane next to Hrd1. This happens when hydrophilic residues become exposed in the transmembrane segment.
Key findings
- Hydrophilic residues trigger degradation
Introducing charged or hydrophilic amino acids into the transmembrane (TM) segment of single-pass proteins was sufficient to trigger Hrd1-dependent degradation. This was demonstrated through cycloheximide-chase experiments in wild-type yeast and strains lacking key ERAD components (Figure 1 in the preprint). The authors used model single-pass proteins containing an idealized hydrophobic TM and compared it to variants in which a single hydrophobic residue was replaced with a charged or hydrophilic one. These modified constructs were rapidly degraded, whereas substitution with hydrophobic residues had no effect.
- Position of hydrophilic residues is critical for degradation rate
Degradation depended strongly on where the hydrophilic residue was located within the TM. Cycloheximide-chase assays showed that residues in the middle or cytosolic side of the membrane triggered the fastest degradation, whereas luminal-proximal positions had weaker effects. To generalize this observation, the authors created a comprehensive library with randomized amino acids at position 10 and analyzed the abundance of each variant using FACS followed by next-generation sequencing. The resulting TM Stability Score correlated closely with the free energy of transferring each amino acid into the lipid phase (Figures 2 and 3 in the preprint).
Mechanistically, the TMs with exposed hydrophilic residues partition into the locally thinned membrane region adjacent to the lateral gate of Hrd1. This movement allows the proteins to escape the energetic penalty of being in the lipid phase by moving into the aqueous environment of Hrd1’s cytosolic cavity. This increased residence time near Hrd1 leads to polyubiquitination.
- Single-pass and multi-pass proteins behave differently
Cycloheximide-chase experiments on model single-pass substrates and endogenous single-pass proteins demonstrated that their degradation requires both Hrd1 and Der1, consistent with their insertion into the thinned membrane region formed cooperatively by these two proteins (Figure 4 in the preprint). In contrast, multi-pass substrates were degraded in an Hrd1-dependent but Der1-independent manner (Figures 5 and 6 in the preprint).
- Hrd1 dimer displaces Der1 to accommodate multi-pass proteins
To explain how multi-pass proteins bypass the need for Der1, the authors determined a cryo-EM structure of a Hrd1 dimer (Figure 6 in the preprint). This revealed that the second Hrd1 monomer occupies the same position normally taken by Der1. Importantly, each Hrd1 subunit contains a large cytosolic cavity, indicating that Der1 must move aside for multi-pass proteins to enter the cavity-adjacent thinned membrane region and undergo degradation.
Future directions and questions for the authors
- The D4-R single-pass construct, which positioned a hydrophilic residue near the luminal leaflet, was surprisingly stable and poorly recognized by the ER-localized ligases. Does this mean that multi-pass proteins also tolerate hydrophilic residues in their luminal regions?
- The authors predict that multi-pass proteins may partition less efficiently and be degraded more slowly than single-pass proteins because they cannot utilize Der1. It would be interesting to quantify this.
- The authors note that Asi and Doa10 recognize substrates via different mechanisms than the partitioning model proposed for Hrd1. It would be interesting to expand on how the other ER-localized ubiquitin ligases identify their substrates.
- Given that the Dsc ligase complex in the Golgi contains components predicted to be structurally similar to Hrd1 and Der1, does the partitioning model also apply to membrane protein quality control mechanisms within the Golgi apparatus?
- To what extent can the partitioning mechanism be extended to higher eukaryotes? Mammalian Hrd1 homologs (gp78, RNF145, Trc8) have distinct substrate specificities. Do the cytosolic cavities of these mammalian homologs provide more specific binding sites that refine the general partitioning mechanism?
doi: https://doi.org/10.1242/prelights.42199
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the molecular biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)