Reduced mitochondrial lipid oxidation leads to fat accumulation in myosteatosis
Posted on: 7 January 2019
Preprint posted on 18 November 2018
Article now published in The FASEB Journal at http://dx.doi.org/10.1096/fj.201802457RR
Why do lipids accumulate following muscle injury? A multi-omics study points to mitochondrial and lipid metabolism dysfunction.
Selected by Pablo Ranea RoblesCategories: biochemistry, pathology, physiology
Introduction
Do you know anyone that had a muscle injury while heavy lifting or practicing sports? Sure you do. Chronic muscle injuries provoke the loss of mobility in the patients, imposing a burden on health care and workers’ compensation systems. Among the muscles usually prone to injury, the rotator cuff is one of the most affected. The rotator cuff is a group of muscles and tendons that stabilize the shoulder joint and let us lift and rotate our arms (Figure 1).
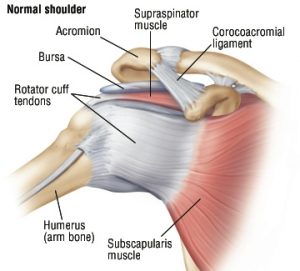
Figure 1. The anatomy of the human rotator cuff. Credit: Harvard Health Publishing
Have you ever wondered what changes happen in that injured muscle after the injury? One of the best characterized effects of muscle injury is pathological lipid accumulation. This is known as myosteatosis (from the Greek words myos-, muscle, steatos-, fat, and -osis, formation). The rotator cuff is particularly susceptible to develop pathological lipid accumulation after injury. Importantly, lipid accumulation in this muscle after injury correlates with a poor outcome after surgical repair (Gladstone et al., 2007). Moreover, recurrence of tears after the injury is quite common in this type of injuries (Isaac et al., 2012). It is known that lipid excess likely impairs muscle regeneration, but the mechanisms driving lipid accumulation in myosteatosis remain largely unknown.
In this study, Gumucio and coworkers used a rat model of rotator cuff injury (Gumucio et al., 2018). The anatomy of the rotator cuff in rats is similar to humans, and this model mimics many of the pathological changes observed in patients with chronic rotator cuff tears (Soslowsky et al., 1996). They studied the supraspinatus muscle, one of the muscles of the rotator cuff (supraspinatus, infraspinatus, teres minor, and subscapularis). Experimental groups were uninjured and injured rats, either 10, 30, or 60 days after the injury. They aimed to characterize the biochemical and cellular pathways that lead to myosteatosis after skeletal muscle injury.
Key findings
They characterized the changes in muscle fiber force production and the biological changes after injury by the integration of different -omics techniques. They evaluated alterations in the rotator cuff transcriptome (by RNA sequencing), proteome, metabolome, and lipidome (using mass spectrometry). The main outcome was the identification of mitochondrial dysfunction and impaired fatty acid oxidation as strong drivers of the pathological steatosis after muscle injury. Then, they studied in detail the hypothesis that mitochondrial dysfunction drives pathological lipid accumulation in torn rotator cuff muscles.
Shotgun lipidomics revealed expected increases in different lipid species at different time points in the injured muscles, such as free fatty acids (FFA), triglycerides, ceramides, sphingomyelins, and some phospholipids. Other lipids displayed a biphasic response, like cholesterol esters, diacylglycerides, and other phospholipids.
The metabolomics data revealed a decrease in nucleoside and nucleotide metabolites, concomitant with an increase in glycolytic and pentose phosphate pathway metabolites. The transcriptomics data showed an induction of well-known pathways in muscle injury, such as autophagy, atrophy, inflammation or fibrosis. It is worth to highlight the decreased mRNA expression of genes involved in mitochondrial function (Krebs cycle and OXPHOS system), lipid uptake and metabolism, and fatty acid oxidation, which was confirmed by proteomics data. Another aspect that appeared in the transcriptomics and proteomics data is oxidative stress. This is deduced due to the increase reactive oxygen species (ROS)-related genes and proteins amount. Moreover, omega-oxidation and peroxisomal metabolism were induced, since transcriptomics data showed augmented expression of Acox1 (Acyl-CoA oxidase 1, an enzyme of peroxisomal beta-oxidation) and Cyp4b1 (from the cytochrome P450 family, related to omega-oxidation of fatty acids). These pathways are usually active when mitochondrial fatty acid oxidation is impaired. These data, together with the increased glycolytic metabolites point to a metabolic shift in injured muscles, from fatty acid oxidation and oxidative phosphorylation to glycolysis.
Finally, a deeper study on mitochondrial function demonstrated that mitochondrial metabolism is impaired in injured muscles, as shown by a reduction in the activity of complexes I, II, and IV, an increase of some antioxidant proteins, and a reduction in the oxidation rate of pyruvate and palmitate. However, mitochondrial content seems to be equal in injured and non-injured muscles, since mitochondrial DNA levels were similar in both groups.
Future directions and questions for authors
Some aspects of this study deserve more attention. For instance, what is the shape of mitochondria in injured muscles? Are they smaller or bigger? Is there peroxisomal proliferation in injured muscles, given the increase of peroxisomal metabolism? Which are the species of acylcarnitines measured? If fatty acid oxidation is impaired, one would expect an increase in some of the acylcarnitines species. Is there autophagy impairment in the injured muscles? The p62 accumulation observed in injured muscles is a classical marker of autophagy impairment. After this study, it would be of interest to know which the next steps are. Can these altered pathways be modulated by drugs? In this way, we would be able to study their effect on muscle regeneration after injury. It would be also of interest to compare human samples from injured muscles with non-injured muscles, to see if these changes are conserved. Finally, one important weakness of this study is that it has been performed only in male rats. Fat content and metabolism is different between men and women, so more studies in female individuals of animal models need to be done to fully understand the pathophysiology of muscle injuries.
What I liked about the study
I liked that the authors integrated different -omics data to gain insights into the molecular physiology of muscle injury. This kind of unbiased approach can shed light on hidden pathological mechanisms. Here, they uncovered a central role of mitochondrial and lipid metabolism in the development of myosteatosis after muscle injury.
References:
Gladstone, J. N., Bishop, J. Y., Lo, I. K. Y. and Flatow, E. L. (2007). Fatty Infiltration and Atrophy of the Rotator Cuff do not Improve after Rotator Cuff Repair and Correlate with Poor Functional Outcome. Am. J. Sports Med. 35, 719–728.
Gumucio, J. P., Qasawa, A. H., Ferrara, P. J., Malik, A. N., Funai, K., McDonagh, B. and Mendias, C. L. (2018). Reduced mitochondrial lipid oxidation leads to fat accumulation in myosteatosis. bioRxiv 471979.
Isaac, C., Gharaibeh, B., Witt, M., Wright, V. J. and Huard, J. (2012). Biologic approaches to enhance rotator cuff healing after injury. J. shoulder Elb. Surg. 21, 181–90.
Soslowsky, L. J., Carpenter, J. E., DeBano, C. M., Banerji, I. and Moalli, M. R. (1996). Development and use of an animal model for investigations on rotator cuff disease. J. shoulder Elb. Surg. 5, 383–92.
doi: https://doi.org/10.1242/prelights.6951
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Also in the pathology category:
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Schistosoma haematobium DNA and Eggs in the Urine Sample of School-Age Children (SAC) in South-West Nigeria
Hala Taha
FUS Mislocalization Rewires a Cortical Gene Network to Drive 2 Cognitive and Behavioral Impairment in ALS
Taylor Stolberg
Also in the physiology category:
Resilience to cardiac aging in Greenland shark Somniosus microcephalus
Theodora Stougiannou
Trade-offs between surviving and thriving: A careful balance of physiological limitations and reproductive effort under thermal stress
Tshepiso Majelantle
Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Muhammed Sinan Malik
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the pathology category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
Also in the physiology category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |











 (No Ratings Yet)
(No Ratings Yet)