DNA-enhanced CuAAC ligand enables live-cell detection of intracellular biomolecules
Posted on: 23 December 2022
Preprint posted on 10 November 2022
It’s all clicking together: @KeqingN and colleagues (@Yifang17957864 and @SRouhanifard) develop a DNA oligomer-conjugated ligand for copper-catalysed azide-alkyne cycloadditions
Selected by Zhang-He GohCategories: molecular biology
Background of the preprint
The copper-catalysed azide-alkyne cycloaddition (CuAAC) is a common strategy to combine an azide-bearing and an alkyne-bearing component. Importantly, this powerful method has a rapid rate of reaction, tolerates aqueous conditions, and is usually free of by-products—making it a bioorthogonal “click” strategy. Despite these advantages, CuAAC has not often been used for intracellular labelling. This is primarily due to copper toxicity, which is responsible for catalysing the generation of reactive oxygen species that are detrimental to the cells’ viability. In this preprint, Nian and colleagues develop a DNA oligomer-conjugated CuAAC accelerating ligand (BTT(1,2)-DNA) and describe how it overcomes several limitations (Figure 1).
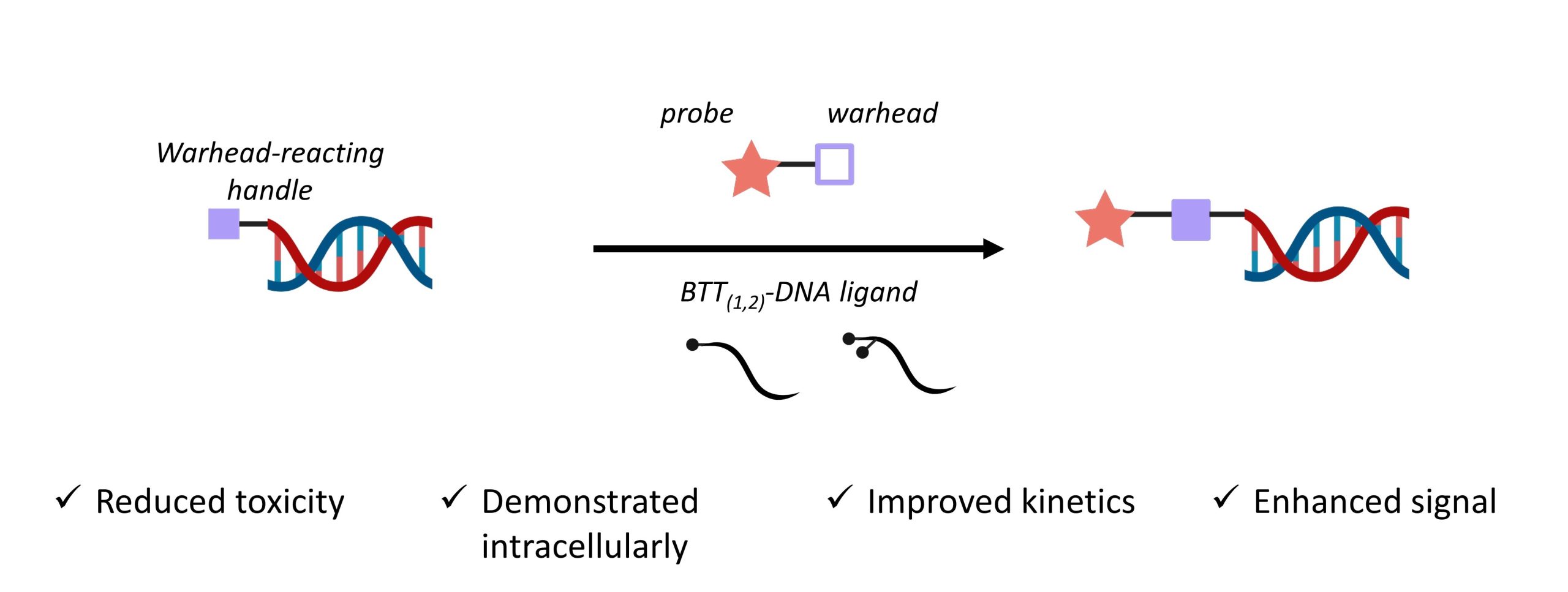
Figure 1. Advantages of the authors’ BTT(1,2)-DNA over current methods.
Key findings of this preprint
Nian and colleagues first synthesised the BTT(1,2)‑DNA ligand, a mixture of DNA-bearing triazole products derived from precursors bearing either one or two azides. Notably, these products were complexed with Cu such that the ratio of Cu:BTT(1,2)‑DNA ligand was 7.5:1 even after dialysis, which is supposed to remove Cu from the ligand. Subsequently, the authors compared the kinetics of the BTT(1,2)‑DNA ligand in CuAAC to that of the non‑DNA bearing BTTAA (complexed with Cu so that the ratio of Cu:BTTAA was 1:2). They showed that BTT(1,2)‑DNA ligand consistently outperformed BTTAA in accelerating the CuAAC reaction for various dyes. Impressively, the Cu-complexed BTT(1,2)‑DNA ligand was able to catalyse the CuAAC at nanomolar ligand concentrations in the absence of free Cu(I).
Next, Nian and colleagues aimed to improve the reaction kinetics by increasing the local concentration of the reaction components. To do so, the authors used DNA splints of various lengths that were complementary to both the target 5’ alkyne DNA and the 3’ BTT(1,2)‑DNA ligand to bring these reaction components into closer proximity to one another. They found that longer splints with more binding sites indeed accelerated the rate of the reaction more.
Finally, the authors demonstrated that their technique could be applied intracellularly. For these experiments, the authors used 5-ethynyl uridine, an alkyne-derivatised uridine analogue that can be metabolically incorporated into RNAs in live cells. In fixed cells, the BTT(1,2)‑DNA ligand improved the fluorescent signal over 3-fold compared to BTTAA. In live HeLa cells, the BTT(1,2)‑DNA ligand was effective in labelling nascent RNAs at concentrations as low as 5 μM. Apart from RNA, the authors also showed that the BTT(1,2)‑DNA ligand was effective in labelling alkyne-bearing cell surface sialic acids. Importantly, because copper-induced cellular toxicity is an important downside in CuAAC, the authors also showed that the BTT(1,2)‑DNA ligand exhibited reduced toxicity in live cells when compared to the commercial BTTAA ligand at the concentrations used in their experiments.
What I like about this preprint
This preprint by Nian and colleagues improves the traditional CuAAC reaction in several ways. First, the new ligand has lower copper-induced cellular toxicity, a common problem associated with CuAAC—this was likely made possible by eliminating the need for free Cu. Doing so has allowed the authors to track the incorporation of a target nucleotide in nascent RNA. Second, this strategy also exhibits improved properties, such as signal intensity and rate, compared to BTTAA when used on dyes.
Future directions
What I find most interesting is the authors’ strategy of using the DNA splint to enhance the CuAAC reaction. Potentially, this would be a powerful method to selectively target certain sequences over others. For instance, to target a particular sequence in cells, it may be possible to use a bespoke DNA splint to bring this sequence into closer proximity to the 3’ BTT(1,2)‑DNA ligand than other sequences. Such a technique would be particularly useful for understanding how DNA is replicated and transcribed into RNA, benefitting fields like genetics and transcriptomics (and likely helping us to understand the epitranscriptome as well).
Questions for the authors
- I find it interesting that the Cu was not dialysed out of the product mixture when purifying the BTT(1,2)‑DNA ligand. Do you think this is representative of the binding strength of the Cu‑BTT(1,2)‑DNA ligand complex? Given that this would also imply a reduced free Cu concentration in aqueous solution, might this also be the reason why your ligand is less toxic than BTTAA?
- Interestingly, the BTT(1,2)‑DNA ligand displays superior labelling properties (e.g. kinetics) over BTTAA, even in the absence of the DNA splint. Why might this be the case—which parts of the structure do you think have enhanced its properties over traditional smaller molecule ligands?
Acknowledgements
Images created using Microsoft Powerpoint and BioRender.
doi: https://doi.org/10.1242/prelights.33434
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the molecular biology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
preLists in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)