Cellular chaining influences biofilm formation and structure in Group A Streptococcus
Posted on: 2 September 2021
Preprint posted on 5 November 2019
Article now published in Biofilm at http://dx.doi.org/10.1016/j.bioflm.2019.100013
Human pathogen Group A Streptococcus (GAS) biofilm is treatment resistant. Biofilm formation and chaining phenotypes differ with varying NaCl concentrations and the presence of lysozymes. A step towards the development of therapeutics?
Selected by SCELSE Summer SchoolCategories: biophysics, microbiology, molecular biology
Names: Jeslyn Poo Shi Ting, Jonas Koh Zhi Xiang, Navin S/O Jeyabalan, Regine Tiong Hui Yi
Background
Group A Streptococcus (GAS), is a human pathogen that causes a variety of human diseases [1,2]. During a GAS infection, the formation of biofilm, which is a complex aggregate of microbes, is an important factor contributing to antibiotic treatment failure [3,4]. It may also play a role in GAS pathogenesis and disease by changing the bacterial transcriptional profile or by modulation of the host response [5-7].
Chaining is a defining morphological feature of GAS, and its contribution to its pathogenicity and biofilm formation has not been deeply investigated. In other bacterial species, chaining contributes to virulence traits like complement evasion and adhesion to host cells, as well as biofilm formation [8-10]. Furthermore, it is poorly understood how GAS regulates its own chain length and it is likely to be complex and highly dependent on environmental conditions.
Previously, a transposon library screen for chain length-influencing genes in S. sanguinis found that a large number of transposon mutants, corresponding to 15% of the genes within the genome, had either a significant increase or decrease in bacterial chain length [11]. This suggests that streptococcal chaining could be a highly regulated process. Growth medium supplemented with homologous antiserum containing anti-M antibodies induces the formation of long chains, while non-immune serum results in chain shortening [12].
In GAS, the autolysin Mur1.2 is predicted to be involved in cell separation and morphology, due to its homology with the peptidoglycan hydrolase LytB in S. pneumoniae [13]. Transcriptomic studies showed that autolysin Mur1.2 is induced by Serine/Threonine kinase Stk, which is an important regulator of cell division, growth and virulence [13]. In addition, the deletion of rgg2, which is a transcriptional regulator, has resulted in overexpression of mur1.2 and increased biofilm formation [14].
In this preprint, the authors investigate how GAS biofilm formation can be directly influenced by host and environmental factors through the modulation of bacterial chain lengths, potentially contributing to persistence and colonization within the host.
Key Findings:
Overview of the experimental design
Bacterial strains used included both GAS and Escherichia coli (E. coli). Growth conditions were adjusted through the supplementation of the growth medium with lysozyme or serum. The Δmur1.2 deletion mutant was constructed by allelic replacement and complementation of the Δmur1.2 knockout strain was performed by cloning the mur1.2 gene to regulate the chaining of GAS cells.
De-chaining of GAS cells is critical for the formation and stability of a dense biofilm
The authors demonstrated that the host environment (presence of lysozyme and NaCl) modulates GAS chain length which regulate GAS biofilm morphology and structure. De-chaining of GAS cells (induced by lysozyme as well as addition of non-immune serum) promotes the formation of a denser and compact 3D structure, improving stability of GAS biofilms (Fig. 1). This point was strongly validated with the use of NaCl (Fig. 2) and deletion of mur1.2 (hyperchaining mutant; Fig. 3), both of which increased chaining of GAS cells, decreased biofilm formation characterised by a loose biofilm, with more fragile architecture. Interestingly, this weaker biofilm has reduced antibiotic tolerance.
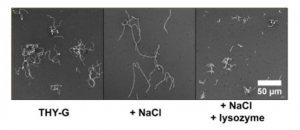
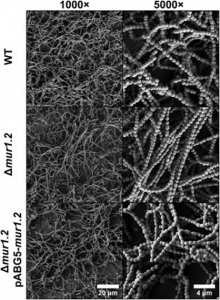
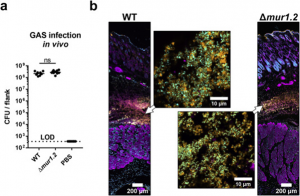
Figure 1. Lysozyme induces de-chaining of GAS cells of four strains, using negative staining (a), crystal violet staining (b) and confocal microscopy (c,d) Adapted from Figure 1 of Matysik et al. (2020).

Figure 2. NaCl causes high chaining morphology of GAS cells which is attenuated by the addition of lysozyme. Adapted from Figure 2a of Matysik et al. (2020).

Figure 3. Deletion of mur1.2 changes the biofilm structure by producing long chains of GAS cells. Adapted from Figure 3 of Matysik et al. (2020).
In vivo analysis of GAS infections on BALB/c mice revealed the host environment influences GAS chaining.
In addition to the in vitro phenotypes observed for GAS de-chaining in the presence of serum or lysozyme, in vivo validation was sought using the mur1.2 mutant on necrotizing fasciitis mice model. Lesions were harvested and colony forming units were enumerated from both mice inoculated with WT GAS and mur1.2 mutants. There were no significant differences in the colony counts 24 hours post-infection and a microscopic evaluation of lesions through immunostaining showed no major differences from chaining. It is shown that in vivo GAS appears in clumps of cells which is a stark difference compared to the in vitro phenotype in absence of both serum or lysozyme. The lack of chaining seen in vivo is highly suggestive of a dominant host derived factor contributing to GAS morphology. Notably these factors could include but are not limited to, protection for GAS from dechaining factors found in serum through phagocytic cell internalisation or cytosolic concentrations of NaCl that could enabling chaining.

Figure 4. GAS in vivo infection in BALB/c mice via subcutaneous injections. Adapted from Figure 5 of Matysik et al. (2020).
What we like about this work:
We like that the authors used a cautious approach to investigate GAS biofilms by applying both in vitro and in vivo models and that they drew on appropriate controls, including de-chaining factors (e.g. lysozyme) in the growth media to simulate a more realistic physiological environment. In addition, the authors opined that classical crystal violet assays may not be the best way to investigate biofilm formation as it does not translate well into in vivo observations. The technical limitations described in this paper will enable future research of GAS biofilm formation and virulence to be more clinically relevant.
Future directions/questions for the authors:
- The authors have tried to induce augmented GAS chaining with NaCl, and have shown that NaCl can reduce biofilm formation. In the future, authors can repeat this experiment to mimic the natural host (in this case human)’s NaCl concentration variations so as to provide clinical relevance.
- It will be interesting for the authors to provide opinions and clinical modelling to show how GAS infection can be treated using this paper’s findings (how GAS can be de-chained, and/or how biofilm formation can be disrupted by inhibiting mur1.2 in vivo).
References:
- Ralph AP, Carapetis JR. 2013. Group a streptococcal diseases and their global burden. Curr Top Microbiol Immunol 368:1–27.
- Ferretti JJ, Stevens DL, Fischetti V, Sims Sanyahumbi A, Colquhoun S, Wyber R, Carapetis JR. 2016. Global Disease Burden of Group A Streptococcus. In Ferretti JJ, Stevens DL, Fischetti VA (ed), Streptococcus pyogenes : Basic Biology to Clinical Manifestations. University of Oklahoma Health Sciences Center, Oklahoma City (OK).
- Baldassarri L, Creti R, Recchia S, Imperi M, Facinelli B, Giovanetti E, Pataracchia M, Alfarone G, Orefici G. 2006. Therapeutic failures of antibiotics used to treat macrolide-susceptible Streptococcus pyogenes infections may be due to biofilm formation. J Clin Microbiol 44:2721–7.
- Conley J, Olson ME, Cook LS, Ceri H, Phan V, Davies HD. 2003. Biofilm formation by group a streptococci: is there a relationship with treatment failure? J Clin Microbiol 41:4043–8.
- Vyas HKN, Proctor EJ, McArthur J, Gorman J, Sanderson-Smith M. 2019. Current understanding of Group A Streptococcal biofilms. Curr Drug Targets doi:10.2174/1389450120666190405095712.
- Ferretti JJ, Stevens DL, Fischetti VAYoung C, Holder RC, Dubois L, Reid SD. 2016. Streptococcus pyogenes Biofilm. In Ferretti JJ, Stevens DL, Fischetti VA (ed), Streptococcus pyogenes: Basic Biology to Clinical Manifestations, Oklahoma City (OK)
- Vajjala A, Biswas D, Tay WH, Hanski E, Kline KA. 2019. Streptolysin-induced endoplasmic reticulum stress promotes group A Streptococcal host-associated biofilm formation and necrotising fasciitis. Cell Microbiol 21:e12956.
- Cho KH, Caparon MG. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Molecular Microbiology 57:1545–1556.
- Dalia AB, Weiser JN. 2011. Minimization of bacterial size allows for complement evasion and is overcome by the agglutinating effect of antibody. Cell Host Microbe 10:486–96.
- Rodriguez JL, Dalia AB, Weiser JN. 2012. Increased chain length promotes pneumococcal adherence and colonization. Infect Immun 80:3454–9.
- Evans K, Stone V, Chen L, Ge X, Xu P. 2014. Systematic study of genes influencing cellular chain length in Streptococcus sanguinis. Microbiology 160:307–15.
- Ekstedt RD, Stollerman GH. 1960. Factors affecting the chain length of Group A Streptococci. Journal of Experimental Medicine 112:687–698.
- Bugrysheva J, Froehlich BJ, Freiberg JA, Scott JR. 2011. Serine/threonine protein kinase Stk is required for virulence, stress response, and penicillin tolerance in Streptococcus pyogenes. Infect Immun 79:4201–9.
- Zutkis AA, Anbalagan S, Chaussee MS, Dmitriev AV. 2014. Inactivation of the Rgg2 transcriptional regulator ablates the virulence of Streptococcus pyogenes. PLoS One 9:e114784.
doi: https://doi.org/10.1242/prelights.30458
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
Also in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
Also in the molecular biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)