Single-cell transcriptome analysis of embryonic and adult endothelial cells allows to rank the hemogenic potential of post-natal endothelium
Posted on: 15 July 2021
Preprint posted on 16 June 2021
Article now published in Scientific Reports at http://dx.doi.org/10.1038/s41598-022-16127-0
Adamov et al. use scRNA-seq to rank the potential of adult mouse tissues to be reprogrammed into HSCs – providing insight into a potential alternative for transplant patients.
Selected by Bobby RanjanCategories: bioinformatics, developmental biology
Background
Every year, thousands of patients require haematopoietic stem cell (HSC) transplantations to cure non-malignant diseases (e.g. thalassemia) and blood cancers like acute lymphocytic leukaemia (Passweg et al., 2019). Unfortunately, these transplantations require a suitable donor, which can be a major limitation. Cellular reprogramming of patient cells into HSCs has therefore been explored as an alternative to circumvent this problem.
In our body, HSCs are produced from endothelial cells, the building blocks of blood vessels, through a process called Endothelial to Haematopoietic Transition (EHT). EHT occurs in the embryo at major haematopoietic sites such as the Aorta-Gonad-Mesonephros (AGM) region and the Yolk Sac (YS). Multiple recent studies in non-human species have shown that non-embryonic/adult endothelial cells could also undergo EHT if exposed to the right conditions.
To explore the EHT potential of adult endothelial cells at single-cell resolution, Adamov et al. selected EHT-related single-cell clusters from three different datasets of the AGM region between E9.5 and E11 in the mouse (Embryo_dataset_1: Hou et al., 2020; Embryo_dataset_2: Vink et al., 2020; and Embryo_dataset_3: Zhu et al., 2020). These cells were integrated with data from 12 adult tissues from Tabula Muris, a consortium of murine single-cell data (Tabula Muris Consortium, 2018).
The authors compared the transcriptomes of adult and embryonic endothelial cells based on gene expression patterns and gene regulatory networks. Based on this analysis, they ranked adult tissues based on the potential of their endothelial cells to undergo EHT.
Key Findings
Single-cell gene and TF expression patterns do not provide direct evidence for EHT in adult mice
To find evidence of EHT in adult tissues, the authors clustered cells in the integrated single-cell dataset and identified fourteen distinct clusters. Of these, only one cluster (of 100% embryonic origin) specifically expressed hemogenic markers such as Runx1 (Sroczynska et al., 2009), Itga4 (Li et al., 2018) and Neurl3 (Hou et al., 2020). No such signature was observed in the adult cells.
Since there was no evidence of EHT in the overall gene expression patterns, the authors specifically looked for the expression of key transcription factors (TFs) crucial to the EHT process and haematopoiesis: Cbfa2t3, Cbfb, Erg, Fli1, Gata1, Gata2, Ldb1, Lmo2, Lyl1, Runx1 and Tal1 (aka “seed TFs”). In particular, the co-expression of Cbfb, Erg, Fli1, Gata2, Lmo2, Lyl1, Runx1 and Tal1 at the single-cell level was characteristic of the endothelial cells initiating the expression of haematopoietic genes (Bergiers et al., 2018). As Runx1 is the main regulator of EHT, (Lancrin et al., 2009; Sroczynska et al., 2009; LieA-Ling et al., 2018) endothelial cells expressing this transcription factor were of particular interest for further investigation.
The abovementioned TFs were highly expressed (detected in nearly all cells) and co-expressed (over half the cells expressed nine out of ten TFs simultaneously) in two out of the three embryonic datasets used. The authors attribute the anomalous TF expression in the third dataset to limitations in the 10X Genomics technology. In adult tissues, TF expression tended to be moderate (detected in roughly 25-50% of cells). The aorta, brain, lung and pancreas tissues had about half of the endothelial cells co-expressing five-to-seven TFs.
Both these results were consistent with the conventional belief in the field that EHT only occurs in embryonic tissues.
While many adult tissues had high overlap of target genes between seed TFs, overlap with Runx1 target genes was rare
The systematic difference in TF expression and co-expression between embryonic and adult endothelial cells could have an effect on their respective regulatory programs. To interrogate these differences in more detail, the authors constructed Gene Regulatory Networks (GRN) using the scTarNet R package (Bergiers et al., 2018). A GRN describes how TFs and their target genes interact with each other to influence gene expression.
The authors used seed TFs to construct a GRN for each tissue, and observed a strong positive association between TFs and their target genes in nearly all adult tissues. In the network constructed from embryonic endothelial cells, Runx1 target genes overlapped with those of Erg and Fli1, which could be indicative of EHT. In adult endothelial cell GRNs, despite the fact that target genes of many seed TFs had strong overlaps, an overlap of Runx1 target genes was rarely observed.
Runx1 gene expression is not sufficient to trigger EHT in adult endothelial cells
Clustering and differential expression analysis of Runx1+ endothelial cells versus the rest of the endothelial cells revealed 13 common marker genes across the adult tissues. Of these, only Cd44, Notch2 and Cd63 have been linked to haematopoiesis (Oatley et al., 2020). Intriguingly, adult Runx1+ endothelial cells showed no evidence of haematopoietic cell fate and expressed none of the key Runx1 targets in EHT (Lacrin et al., 2012), indicating that expression of Runx1 is not sufficient for EHT in adult endothelial cells.
Pancreas, brain, kidney and liver are the most suitable for reprogramming
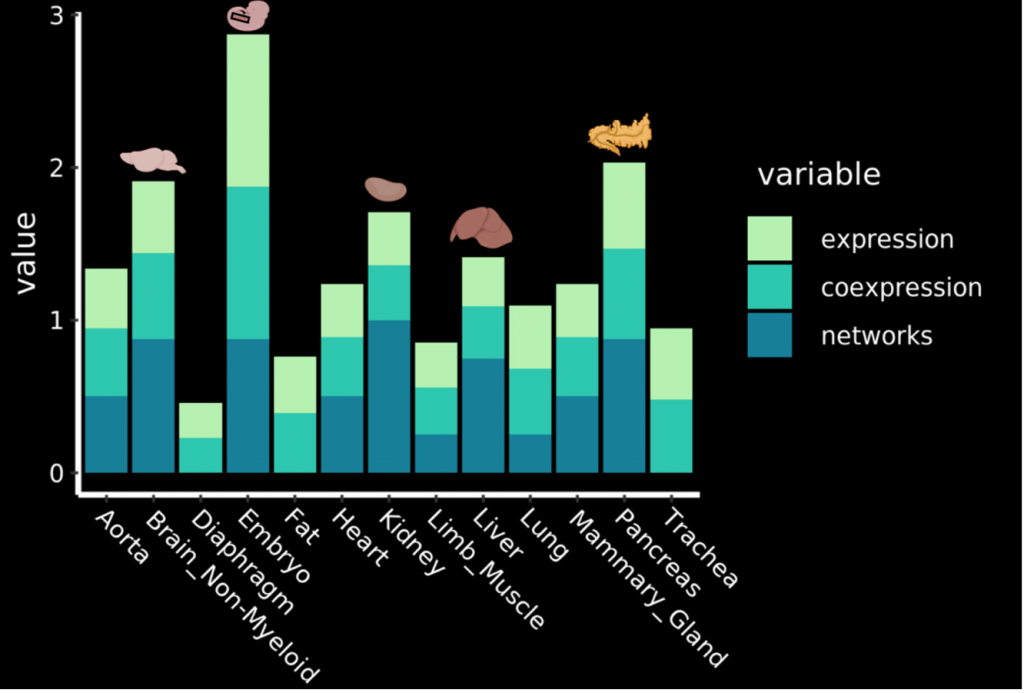
Using the analyses conducted above, the authors computed a normalized aggregate score to rank adult tissues based on their likelihood to undergo EHT based on the following criteria:
- Expression of seed TFs,
- Co-expression of seed TFs, and
- Overlap between target genes of seed TFs in endothelial cells.
Figure 1 shows the relative aggregate scores of the adult tissues. In summary, the pancreas, brain, kidney and liver are the four most promising candidate tissues for undergoing EHT.
Why I chose this preprint
As described above, there is significant clinical value in reprogramming cells from adult tissues into HSCs for patients requiring transplantations. In this preprint, the authors attempt to identify adult tissues having the highest potential for cellular reprogramming into HSCs using a computational single-cell-based strategy. In the process, they also make novel biological discoveries regarding the role of Runx1 in EHT. This resource could be a stepping-stone for attempting in vivo reprogramming by targeting the most promising endothelial cells in the adult organism.
References
- Passweg JR, Baldomero H, Basak GW, et al. The EBMT activity survey report 2017: a focus on allogeneic HCT for nonmalignant indications and on the use of non-HCT cell therapies. Bone Marrow Transplant. 2019 Oct;54(10):1575-1585. doi: 10.1038/s41409-019- 0465-9.
- Hou S, Li Z, Zheng X, et al. Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res. 2020 May;30(5):376-392. doi: 10.1038/s41422-020-0300-2.
- Lancrin C, Mazan M, Stefanska M, et al. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012 Jul 12;120(2):314-22. doi: 10.1182/blood-2011-10-386094.
- Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018 Oct;562(7727):367-372. doi: 10.1038/s41586-018-0590-4.
- Vink CS, Calero-Nieto FJ, Wang X, et al. Iterative Single-Cell Analyses Define the Transcriptome of the First Functional Hematopoietic Stem Cells. Cell Rep. 2020 May 12;31(6):107627. doi: 10.1016/j.celrep.2020.107627.
- Zhu Q, Gao P, Tober J, et al. Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood. 2020 Aug 13;136(7):845-856. doi: 10.1182/blood.2020004801.
- Oatley M, Vargel Bölükbası Ö, Svensson V, et al. Single-cell transcriptomics identifies CD44 as a marker and regulator of endothelial to haematopoietic transition. Nat Commun. 2020 Jan 29;11(1):586. doi: 10.1038/s41467-019-14171-5.
- Bergiers I, Andrews T, Vargel Bölükbaşı Ö, et al. Single-cell transcriptomics reveals a new dynamical function of transcription factors during embryonic hematopoiesis. Elife. 2018 Mar 20;7:e29312. doi: 10.7554/eLife.29312.
- Li D, Xue W, Li M et al. VCAM-1+ macrophages guide the homing of HSPCs to a vascular niche. Nature. 2018 Dec;564(7734):119-124. doi: 10.1038/s41586-018-0709-7.
- Sroczynska P, Lancrin C, Kouskoff V, Lacaud G. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood. 2009 Dec 17;114(26):5279-89. doi: 10.1182/blood-2009-05-222307.
- Lie-A-Ling M, Marinopoulou E, Lilly AJ, et al. Regulation of RUNX1 dosage is crucial for efficient blood formation from hemogenic endothelium. Development. 2018 Mar 12;145(5):dev149419. doi: 10.1242/dev.149419.
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009 Feb 12;457(7231):892-5. doi: 10.1038/nature07679.
doi: https://doi.org/10.1242/prelights.30062
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the developmental biology category:
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)