Discovery and visualization of uncharacterized drug-protein adducts using mass spectrometry
Posted on: 10 July 2021
Preprint posted on 25 June 2021
Article now published in Analytical Chemistry at http://dx.doi.org/10.1021/acs.analchem.1c04101
It all adds up: using mass spectrometry to identify, understand, and characterise drug-protein adducts
Selected by Zhang-He GohCategories: biochemistry, pharmacology and toxicology
Background of the preprint
Every day, we are exposed to many chemicals daily in the food we eat and the air we breathe. Often this exposure occurs in the form of pollution, but it can also arise when we take medication for the treatment of various diseases. These chemicals from extraneous sources are known as xenobiotics.
In our body, proteins fulfil important structural and functional roles. Unfortunately, they can react with these xenobiotics. The products that result from these reactions between xenobiotics and proteins—known as protein adducts—are responsible for various toxic effects. Sometimes these can manifest as allergies, which is why many medicines have adverse drug reactions. Evidently, modulating the formation of these adducts is important for human health: if we can modify their underlying mechanisms, we would be able to reduce the negative consequences of adduct formation, such as medicine side effects.
However, to date the identification and characterisation of these protein-chemical adducts remains a major challenge. Existing methods focus on chromatography, immunological techniques, and mass spectrometry to detect these adducts. However, these methods have two key limitations: they are not sufficiently sensitive, and often need to be built on a prior understanding of the chemical composition of these adducts, yet most adducts have not been characterised.
In this work, Riffle et al. describe the application of Magnum and Limelight, two new tools to study protein-drug adducts. Magnum is a customised adduct discovery algorithm and Limelight is a web-based analysis platform. Using CYP3A4, a metabolic enzyme responsible for oxidation, as well as raloxifene, the authors establish a workflow to show how Magum and Limelight can be used to characterise these adducts without the need for a prior understanding of the adducts formed.
Key findings of this preprint
(A) Magnum and Limelight
Riffle et al. developed Magnum, a ‘xenobiotic-protein adduct discovery algorithm’, to analyse tandem mass spectrometry (MS) (MS-MS) spectra. MS-MS is a highly sensitive and specific technique that offers significant advantages over single quadrupole MS. By involving multiple mass analysers, MS-MS is particularly well-suited to analyse biomolecules like proteins.
The authors highlight four features of Magnum that are key to its advantages compared to existing methods. First, Magnum analyses data pertaining to adduct-modified MS-MS ion series that allows better identification of adduct localisation, i.e. the site of adduct formation on the protein. Second, Magnum can restrict adduct localisation to specific amino acids, allowing chemical intuition to guide the search. Third, Magnum detects diagnostic reporter ions that are found in characteristic adduct fragmentation patterns. Fourth, Magnum differentiates between user-defined and undefined modifications. This feature reduces the chance of generating false positive identifications, which are more likely to occur during undefined searches.
Three criteria were used to assess Magnum: precision, recall, and accuracy. Precision was defined as the fraction of all answers that are correct; Magnum’s precision exceeded 0.9 when set at a 1% false discovery rate (set to minimise the rate of false positives generated by the algorithm). Furthermore, Magnum also exhibited a good recall, defined as the fraction of total correct answers identified, as well as a high accuracy of monoisotopic mass. Because many pharmaceutical drugs contain halogen isotopes like bromine and chlorine, the authors included code to enhance Magnum’s ability to correctly identify their respective adducts, a feature that is critical for their identification and subsequent characterisation.
To enable easier analysis of adduct data, Riffle et al. next developed Limelight, a web application to analyse, visualise, and share MS proteomics data.
(B) Identification of novel raloxifene adducts in CYP3A4 and P450-reductase
CYP3A4 is an enzyme that is responsible for the oxidation of many xenobiotics and requires P450-reductase to perform catalysis. CYP3A4 also oxidises raloxifene, producing several electrophilic species. A single 471 Da adduct on cysteine 239 has previously been identified.
Intriguingly, using Magnum and Limelight, Riffle et al. found 471 Da adducts not only on cysteine, but also on tryptophan and tyrosine, suggesting that raloxifene also reacts with these nucleophilic residues. Based on these data, the authors discuss the formation of adducts between raloxifene’s metabolites and CYP3A4, observations that may hint at key positions in the protein related to metabolite transport. These findings may in turn lead to important conclusions on the overall structure and function of CYP3A4 as well as its interactions with various ligands.
What I like about this preprint
I selected this preprint because of its potential impact on chemical biology, as well as its significance to related fields like medicine and toxicology (Figure 1). First, by identifying site-specific adducts based on MS spectra, this method may be extremely useful for proteomic applications, in which identifying adducts at selected sites have important downstream implications. For instance, the formation of these adducts could point to a particular residue’s reactivity, allowing for the development of methods to improve selectivity that would enable researchers to either selectively label (or avoid labelling) it. Alternatively, they may also shed light on the accessibility of the site. If the adduct is formed at a given amino acid residue, the amino acid residue could be in an important binding pocket, or perhaps in a surface-exposed environment. The upshot: a paradigm shift in how researchers identify druggable sites on proteins that could aid in drug development.
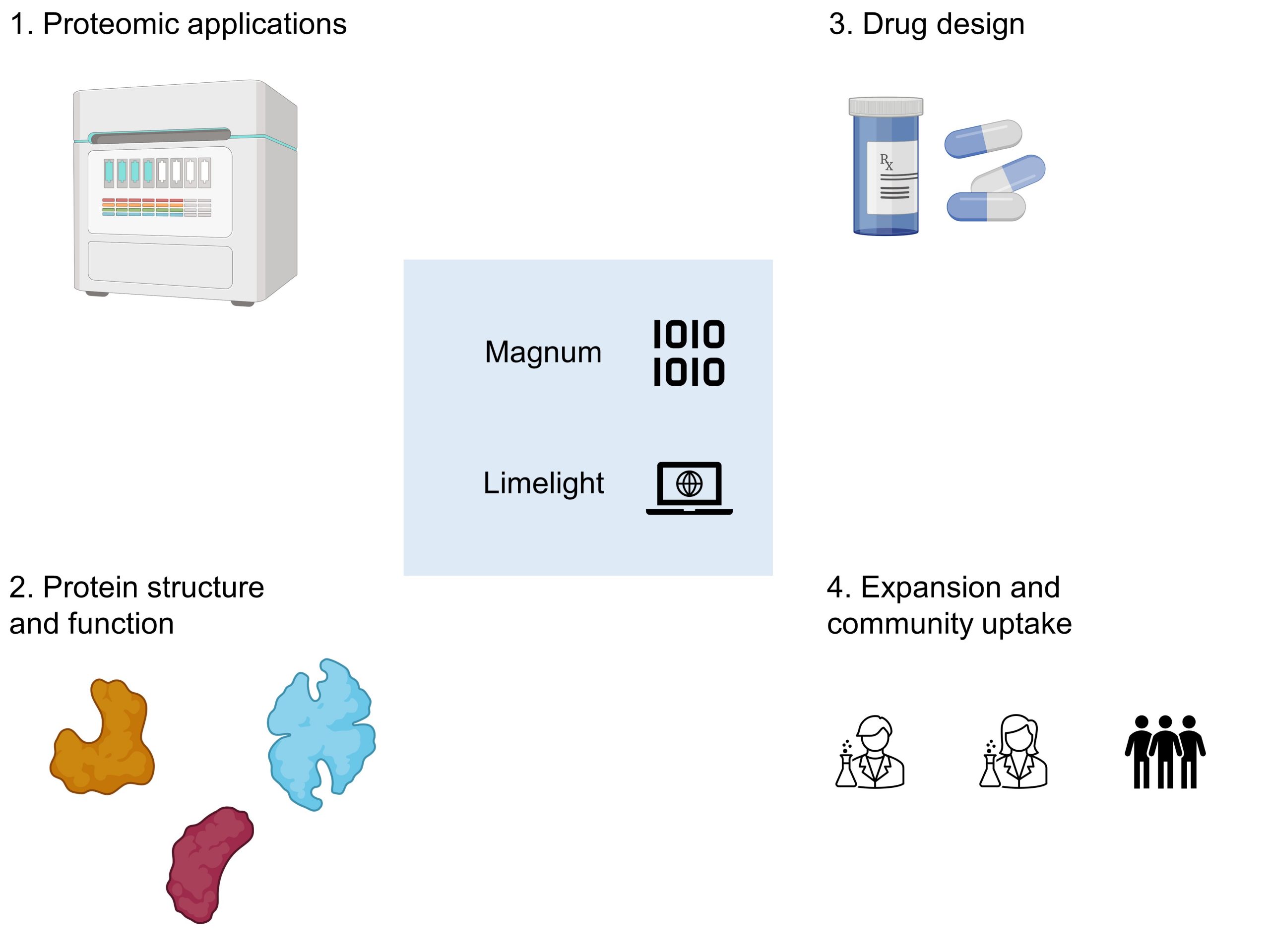
Figure 1. Significance of this work by Riffle et al.
Second, as Riffle et al. discuss in their study, understanding where adducts are formed could help to elucidate the structure and function of proteins. In their work, the authors cite substrate access and metabolite egress channels for this application. MS studies could help to provide evidence for hypotheses in cases where understanding the mechanism could be experimentally challenging.
Third, understanding the mechanism of formation of these adducts could enable improved drug design. While chemically reactive groups are well-understood in drug design, their metabolism in the body is less well defined. Indeed, the ability to understand and predict a drug’s metabolism, a significant contributor to drug toxicity, has been a major challenge in the field of pharmacokinetics for decades. While a few general mechanisms for adverse drug reactions arising from protein-xenobiotic adduct formation have been described, they are still mostly idiosyncratic. Thus, by profiling the reactivity of drugs’ metabolites with functionally important proteins in the body, this technology could enable the development of better medicines with fewer side effects.
Future directions
In addition to the scientific branches of innovation that I have highlighted above, I posit two other ways in which this interesting tool may move the field forward. The first pertains to the Limelight package itself: it is open source and has been made freely available on Github, and collaborations may be possible for further improvements that could be built on its existing capabilities. Given the interdisciplinary nature of research, in which scientists are increasingly encouraged to venture beyond their field of expertise, Limelight could be useful to a wider scientific audience. Researchers from different backgrounds, including computational chemists, chemical biologists, and medicinal chemists, could offer suggestions to improve Limelight’s capabilities. This would transform it into a more generalised platform for different applications.
Related to collaborations are considerations pertaining to usability, such as user design and user experience. These are important features of any application, even those developed for the scientific world. In this regard, researchers from different backgrounds could give feedback or even contribute towards Limelight’s design, as that will likely accelerate its use by the wider scientific community.
Acknowledgements
All images created using Microsoft Powerpoint and BioRender.
Questions for authors
- It is interesting that these methods are suitable for the characterisation of adducts formed from proteins, whether they react with metabolites or with inherently reactive xenobiotics, even without input pertaining to prior knowledge of adduct formation. How does Magnum differentiate between adduct formation at a given amino acid residue over another? In the CYP450 example provided in the preprint, for instance, how does Magnum identify adducts that have undergone addition at cysteine versus tryptophan or tyrosine, as they are likely to have the same mass?
- The adducts formed are likely to be stoichiometric, in that mono-adducts are formed between protein and metabolite in a 1:1 ratio, bi-adducts are formed between protein and metabolite in a 1:2 ratio, etc.. Given that MS-MS is a quantitative method for analysis, did you also observe these ratios between the reporter fragment ions (that arise from the adduct) and the other peptide fragments in the spectra?
doi: https://doi.org/10.1242/prelights.29984
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Also in the pharmacology and toxicology category:
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
In vitro pharmacokinetics and pharmacodynamics of the diarylquinoline TBAJ-587 and its metabolites against Mycobacterium tuberculosis
Zhang-He Goh
Beyond venomous fangs: Uloboridae spiders have lost their venom but not their toxicity
Daniel Fernando Reyes Enríquez, Marcus Oliveira
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the pharmacology and toxicology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Drug use in special populations
Any drugs that are being used in special populations: Patients with liver and kidney failure, in paediatrics, in geriatrics, and in pregnant or lactating patients. Includes the discovery of factors that could potentially affect drug use in these special populations.
| List by | Zhang-He Goh |
Toxicology of toxicants, existing therapeutics, and investigational drugs
Preprints that describe the toxicology of environmental pollutants and existing and upcoming drugs. Includes both toxicokinetics and toxicodynamics, as well as technological improvements that will help in the characterisation of this field.
| List by | Zhang-He Goh |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (1 votes)
(1 votes)