In situ differentiation of iridophore crystallotypes underlies zebrafish stripe patterning
Posted on: 15 April 2020
Preprint posted on 25 March 2020
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-020-20088-1
How zebrafish get their stripes – a new model of how iridophores pattern the skin
Selected by Hannah BrunsdonCategories: developmental biology
Background
The famous zebrafish stripes have often been used as a model for pigment patterning in animals. Three pigment cell types contribute to the stripe pattern – black melanophores in the stripes themselves, yellow xanthophores in the interstripe regions, and reflective iridophores which reside in both. Iridophores have different morphologies depending on their local environment – they are cuboidal and densely packed within interstripes, whereas within stripes they are more stellate and spaced out.
Previous work[1] investigating how the zebrafish stripe pattern emerges and propagates has suggested that iridophores take a central role in organising the process. The current model is that iridophores proliferate, differentiate and pack together densely to form the first interstripe in juvenile fish. Over time, some of these interstripe iridophores adopt a looser morphology and migrate into developing stripes. After a period of proliferation, some of these ‘loose’ iridophores then reaggregate again to form a second interstripe. Therefore, a single iridophore cell type is predicted to transition between dense and loose configurations to pattern the fish.
However, this preprint by Gur, Bain and colleagues proposes an alternative model, whereby iridophore progenitors do not interconvert between dispersed and dense morphologies, but instead take cues from surrounding melanophores to differentiate into one of two iridophore subtypes – stripe and interstripe – with distinct physical and transcriptomic characteristics.
Key findings
In order to study the movements of iridophores during patterning, the team conducted live imaging experiments using the iridophore-specific fluorescent reporter line Tg(pnp4a:mCherry). Despite imaging larvae for over 300(!) hours, the authors did not observe any interconversion of pnp4a:mCherry+ iridophores between interstripes and stripes, conflicting with the previous migration model. To test this further, the team conducted a fate mapping experiment using a pnp4a:mCherry reporter line also containing nuclear mEos, a photoconvertible green to magenta fluorescent protein. A week after photoconversion of an interstripe region, pnp4a:mCherry cells had white nuclei, from a mixture of photoconverted magenta, and more recently translated green unconverted mEos fluorescence. In stripes however, only pnp4a:mCherry cells with green nuclei were observed, confirming that no interstripe iridophores migrated into this region.
This led the authors to hypothesise that rather than being from the same differentiated cell population, stripe and interstripe iridophores might represent distinct cell subtypes. To test this, they investigated the physical and transcriptional differences between the two in greater detail.
Iridophores are reflective because they contain stacks of crystalline guanine. Using cryogenic scanning electron microscopy and synchrotron-based micro X-ray diffraction, the authors showed that stripe iridophores contained regular, neatly-aligned guanine crystals. In contrast, guanine crystals in interstripe iridophores were highly disordered (see part of preprint Figure 3 below).
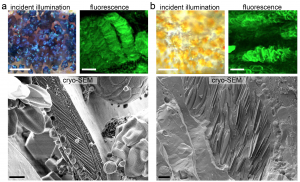
From preprint Figure 3 (with permission from authors). Left – blue stripe iridophores and black melanophores under incident illumination and under fluorescence after staining with malachite green. Underneath, cryo-SEM images show guanine crystal stack architecture. Right – as left, but for interstripe iridophores and yellow xanthophores.
So what might influence which iridophore subtype progenitors become? The team hypothesised that the surrounding environment, in particular stripe melanophores, may influence iridophore crystallotype. They compared the X-ray diffraction patterns in skin of mitfa genetic mutants, which completely lack melanophores, to albino mutants, in which melanophores are present but lack melanin. This revealed that the vast majority of iridophores were of the interstripe disordered crystallotype in mitfa mutant fish. However, in albino fish, ordered stripe iridophore crystallotypes were present within stripes of unpigmented melanophores, alternating with disordered interstripe crystallotypes. Additionally, they used a temperature sensitive allele of mitfa to conditionally add or remove melanophores, and observed that iridophores differentiated into the ordered crystallotype only when melanophores were present.
Taken together, this suggests that stripe and interstripe iridophores arise by in situ differentiation depending on their surrounding environment, and represent distinct cellular crystallotypes.
Why I chose this preprint
I must admit that in these unusual times I found the pretty images of zebrafish stripes in this preprint quite soothing! It was also good to learn more about iridophores, which aren’t nearly as well covered in the literature as melanophores for example. More specifically, I liked how the live imaging, fate mapping analyses, and different transgenic lines were used in conjunction with X-ray diffraction and SEM to investigate the guanine crystal structure – something I’ve not seen in zebrafish papers before but really neatly illustrates the differences between stripe and interstripe iridophores.
Questions for the authors
- Previous work supporting the ‘morphogenic respecification’ model used a sox10-based fluorescent reporter line for tracing analysis, and images these cells migrating and proliferating in stripes and interstripes[1]. Why do you think your findings with the pnp4a reporter line are different? Might sox10+ iridophores represent an earlier iridophore progenitor still able to migrate, and pnp4a expression only occur after migration ends?
- Connected to this, how specific to iridophores is pnp4a? Cell clusters other than iridophores were detected by the RNA-seq analysis, and also express high levels of pteridine and carotenoid-associated genes – could these be xanthophores? Or other cell types entirely?
- Do you have any idea of the mechanism whereby melanophores might influence iridophore subtype? Do you think xanthophores might also influence iridophore morphology in a similar way?
References
- Singh, A.P., Schach, U., & Nusslein-Volhard, Proliferation, dispersal and patterned aggregation of iridophores in the skin prefigure striped colouration of zebrafish. Nat. Cell. Biol. 16, 607-614
- Spiewak, J. E. et al. Evolution of Endothelin signaling and diversification of adult pigment pattern in Danio fishes. PLoS Genet 14, e1007538, doi:10.1371/journal.pgen.1007538 (2018).
- Saunders, L. M. et al. Thyroid hormone regulates distinct paths to maturation in pigment cell lineages. eLife 8, doi:10.7554/eLife.45181 (2019).
- Frohnhofer, H. G., Krauss, J., Maischein, H. M. & Nusslein-Volhard, C. Iridophores and their interactions with other chromatophores are required for stripe formation in zebrafish. Development 140, 2997-3007, doi:10.1242/dev.096719 (2013).
- Patterson, L. B. & Parichy, D. M. Interactions with iridophores and the tissue environment required for patterning melanophores and xanthophores during zebrafish adult pigment stripe formation. PLoS Genet 9, e1003561, doi:10.1371/journal.pgen.1003561 (2013).
- Mahalwar, P., Singh, A. P., Fadeev, A., Nusslein-Volhard, C. & Irion, U. Heterotypic interactions regulate cell shape and density during color pattern formation in zebrafish. Biology open 5, 1680-1690, doi:10.1242/bio.022251 (2016).
doi: https://doi.org/10.1242/prelights.18558
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Cooperation between cortical and cytoplasmic forces shapes planar 4-cell stage embryos
Corentin Mollier, Shivani Dharmadhikari
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)