LincRNAs enable germ cells differentiation by promoting PUF proteins condensation
Posted on: 30 October 2023
Preprint posted on 27 August 2023
The root of stemness - lincRNAs promote FBF-2 condensation to prevent continuous mitosis in the germline.
Selected by Umutcan Kaan Bozan, Chee Kiang Ewe, preLights peer supportCategories: developmental biology
Background:
Proliferative germline stem cells can differentiate into gametes, which are essential for reproduction (Li & Xie, 2005). The overall organization of the C. elegans germline is similar to that in other organisms, containing self-renewing stem cells at the distal end and mature gametes at the proximal end, with germ cells at various stages of differentiation in between. The distal germline can generally be divided into three zones: early progenitor zone (early PZ), late progenitor zone (late PZ) and leptotene/zygotene zone (LZ). The stem cell niche is formed and kept at the early PZ by the distal tip cell (DTC) through the activation of the conserved GLP-1/Notch signaling pathway. The early PZ consists of stem cells, while the late PZ contains stem cells as well as the meiotic S-phase cells. As the cells enter the LZ, they enter the first stages of meiosis, undergoing a proliferation-to-differentiation transition (Albert & Schedl, 2019) (Figure 1).
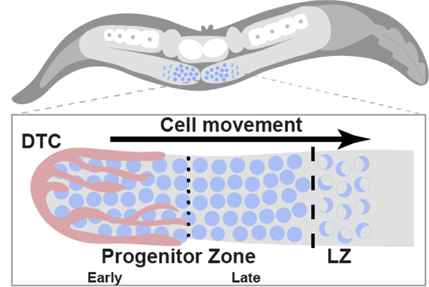
Figure 1: C. elegans germline organization. Taken from Falk et al. Figure 1.
Two members of the conserved PUF family of RNA-binding proteins, FBF-1 and 2, are the major regulators of germline stem cell development in C. elegans. The FBFs primarily repress gene expression in the germline by destabilizing target transcripts or blocking translation. Among FBF targets are cell cycle regulators and several long intervening ncRNAs (lincRNAs) (Nishanth & Simon, 2019). Inactivating fbf-1 or fbf-2 alone does not significantly affect fertility; however, the double mutant is sterile because of a disruption in stem cell maintenance, demonstrating the crucial roles of the PUF proteins in regulating stem cell fate (Crittenden et al., 2002).
Although FBFs share many targets and perform overlapping functions, they have opposing effects on germline stem cell dynamics. FBF-1 facilitates meiotic mRNA degradation and/or transport from the stem cell region, whereas FBF-2 primarily inhibits translation (Voronina et al., 2012). It was previously proposed that the FBFs are expressed in a gradient such that high FBF levels in the early PZ promote a “mitotic” or undifferentiated cell fate and that their expression drop in late PZ and LZ relieves the repression of meiosis-promoting mRNAs (Byrd & Kimble, 2009). Curiously, however, FBF proteins are found in late PZ and LZ (Chen et al., 2012) (see more below), suggesting that their activity, rather than expression, is inhibited at these stages through a yet unknown mechanism that enables germ cell differentiation.
In this preprint, Falk and colleagues could show that three lincRNAs, linc-4, linc-29, and linc-7, which are known to bind to FBF-1 and -2, control meiotic entry and promote germ cell differentiation by sequestering FBFs in germline P granules (perinuclear RNA-protein condensates), thereby blocking their pro-mitotic functions.
Key Findings:
- FBF-1 & FBF-2 are expressed in early meiotic prophase.
In contrast to the “gradient model” which posits that differential expression of FBF proteins controls the mitosis/miosis decision (Byrd & Kimble, 2009), this preprint and a previous study (Chen et al., 2012) found that the FBFs are present throughout the distal germline, with the highest expression in the PZ. While FBF-1 levels drop modestly in the LZ, FBF-2 levels remain high in all zones. Hence, the authors hypothesized that FBFs are suppressed in cells undergoing miosis, perhaps by long non-coding RNAs, which have previously been shown to inhibit the actions of PUF proteins in mammals.
- Three lincRNAs interact with FBFs and inhibit their pro-mitotic functions.
A previous study identified six lincRNAs that are highly expressed in the C. elegans germline. Inactivating individual lincRNAs did not cause any obvious fertility defects (Ishtayeh et al., 2020). Three of the lincRNAs, linc-4, linc-7, and linc-29, have previously been shown to bind the FBFs (Prasad et al., 2016). In this study, the authors found that deleting either linc-4 or linc-7 alone caused a very modest reduction in brood size, while removing both lincRNAs simultaneously led to a significant decrease in fertility. Similarly, a linc-29 single mutant showed only a small drop in brood size whereas the linc-4; linc-29; linc-7 triple deletion mutant (3xlnc) had the smallest brood size compared to WT and the double mutants.
To test whether the FBF-binding ability of lincRNAs is important for fertility, the authors deleted the FBF binding element (FBE) of linc-7. Removing FBE in linc-7 in a linc-4; linc-29 mutant background significantly reduced brood size, phenocopying 3xInc. Together with the findings that linc-4 and -7 are upregulated in meiotic germ cells, the authors put forth a model in which the binding of lincRNAs to FBFs suppresses their activity, promoting meiosis.
- lincRNAs antagonize FBF-2, but not FBF-1, to promote meiosis.
By examining the number of nuclei and mitotic cells in the PZ, the authors found that 3xlnc had a similar effect as fbf-1(-) such that both showed a smaller number of PZ nuclei and a higher mitotic index than WT. Removing fbf-1 in the 3xlnc mutant further exacerbated this phenotype. On the other hand, removing fbf-2 led to an increase in PZ nuclei. Furthermore, inactivating the three lincRNAs in fbf-2(-) did not enhance its phenotype, suggesting that linc-4, linc-29, and linc-7 are epistatic to FBF-2.
Next, the authors performed RNA-seq analysis on WT, the fbf-2(-) and the 3xlnc mutants. Strikingly, they found that 102 of the 221 downregulated genes in 3xlnc physically interact with FBF-2, suggesting that the three lincRNAs normally antagonize the repressive effects of FBF-2 on germline genes.
- FBF-2 condensation paves the way to meiosis.
Next, the authors investigated the cellular localization of the lincRNAs using smFISH, focusing on linc-7. They observed expression of linc-7 in perinuclear P granules and nucleoplasm. They also noticed that the signal increased and became more perinuclear as the germ cells entered the LZ, where meiosis starts. Additionally, the authors found that linc-7 almost exclusively co-localized with an FBF-2 reporter in the perinuclear condensate, especially in the germ cells in the late PZ and LZ.
Finally, the authors tested whether the lincRNAs are important for FBF-2 condensation. Indeed, removing linc-4, -29, and -7, led to a reduction of FBF-2 perinuclear condensates; instead, FBF-2 localized to the cytoplasm. Together, these results suggest that lincRNAs sequester FBF-2 in the P granules, thereby promoting entry to meiosis and germ cell differentiation.
What I liked about this preprint:
This preprint closes the gap on how exactly the entrance to meiosis occurs in the C. elegans germline. I liked this preprint because I think that non-coding RNAs have many undiscovered roles during development as elucidated here. It was clever to think that proteins like FBFs, which can bind RNAs, can also be regulated by (nc)RNAs.
Questions to the authors:
- As shown in supplementary figure 2, the linc-29 expression decreases towards LZ, which is not the case with linc-4 and linc-7, yet its deletion causes less fertile worms. Could linc-29 have another mechanism of action instead of decreasing FBF-2 condensation? Have you also checked the single mutants’ levels of FBF-2 condensation?
- What might regulate lincRNA levels throughout the germline?
- Do FBFs have any roles in downstream gamete differentiation? Why are they inhibited rather than degraded?
References
Albert, J., & Schedl, T. (2019). Biology of the Caenorhabditis elegans Germline Stem Cell System. Genetics, 213(4), 1145–1188. https://doi.org/10.1534/genetics.119.300238
Byrd, D. T., & Kimble, J. (2009). Scratching the niche that controls Caenorhabditis elegans germline stem cells. Seminars in Cell & Developmental Biology, 20(9), 1107–1113. https://doi.org/10.1016/j.semcdb.2009.09.005
Chen, D., Zheng, W., Ai, L., Uyhazi, K. E., Zhao, H., & Lin, H. (2012). Pumilio 1 Suppresses Multiple Activators of p53 to Safeguard Spermatogenesis. Current Biology, 22(5), 420–425. https://doi.org/10.1016/j.cub.2012.01.039
Crittenden, S. L., Bernstein, D., Bachorik, J. L., Thompson, B. E., Gallegos, M., Petcherski, A. G., Moulder, G., Barstead, R., Wickens, M., & Kimble, J. (2002). A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature, 417(6889), 660–663. https://doi.org/10.1038/nature754
Ishtayeh, H., Achache, H., Kroizer, E., Rappaport, Y., Itskovits, E., Gingold, H., Best, C., Rechavi, O., & Tzur, Y. B. (2020). Systematic analysis of long intergenic non-coding RNAs in C. elegans germline uncovers roles in somatic growth. RNA Biology, 18(3), 435–445. https://doi.org/10.1080/15476286.2020.1814549
Li, L., & Xie, T. (2005). STEM CELL NICHE: Structure and Function. Annual Review of Cell and Developmental Biology, 21(1), 605–631. https://doi.org/10.1146/annurev.cellbio.21.012704.131525
Nishanth, M. J., & Simon, B. (2019). Functions, mechanisms and regulation of Pumilio/Puf family RNA binding proteins: a comprehensive review. Molecular Biology Reports, 47(1), 785–807. https://doi.org/10.1007/s11033-019-05142-6
Prasad, A., Porter, D. F., Kroll-Conner, P., Mohanty, I., Ryan, A. R., Crittenden, S. L., Wickens, M., & Kimble, J. (2016). The PUF binding landscape in metazoan germ cells. RNA, 22(7), 1026–1043. https://doi.org/10.1261/rna.055871.116
Voronina, E., Paix, A., & Seydoux, G. (2012). The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development, 139(20), 3732–3740. https://doi.org/10.1242/dev.083980
doi: https://doi.org/10.1242/prelights.35857
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)