Nutrient-regulated dynamics of chondroprogenitors in the postnatal murine growth plate
Posted on: 23 February 2023 , updated on: 27 February 2023
Preprint posted on 21 January 2023
Fasted bones grow fast later: chondroprogenitors in the growth plate of murine long bones adapt to dietary restriction, leading to catch-up growth during refeeding.
Selected by Alberto Rosello-Diez, Boya (Hannah) Zhang, Chee Ho H'ngCategories: developmental biology
Background
One of the most fascinating characteristics of growing animals is that, after a transient developmental perturbation, they show a tendency to regain a normal growth trajectory. This phenomenon is referred to as ‘catch-up growth’, defined by Prader and Tanner (1) as an example of canalization (robustness in the phenotypic traits in response to any perturbations), a concept coined by Waddington (2) and independently proposed by Schmalhausen (3). While some systemic (neuroendocrine) mechanisms were initially proposed to explain catch-up growth (4), the observation that it can independently affect specific organs or tissues shifted the focus towards local mechanisms (5).
The long bones within the limbs have been well studied in this regard. Their growth takes place via endochondral ossification, whereby a transient cartilage template is first formed and progressively replaced by bone (6). The primary ossification center in the shaft of the bone soon divides the initial cartilage into two growing units at both bone ends, called growth plates (Figure 1). Cells in the cartilage (chondrocytes) transition, from the ends towards the center, through subsequent differentiation states: chondroprogenitors in the resting zone are progressively recruited into the proliferative pool of flat chondrocytes, arrayed in longitudinal columns, which subsequently enlarge and differentiate towards hypertrophic chondrocytes (Figure 1). These cells either die or transdifferentiate to bone-laying cells (osteoblasts), so that the cartilage matrix is replaced by bone, adding length to both ends of the bone shaft.
In order to sustain growth over an extended period of time, this system must find a delicate balance between production of cartilage on one end of the growth plate and destruction on the other. Therefore, chondroprogenitors have been a subject of active research for the last 30 years or so, with a focus on their ability to self-renew and the limit (or lack thereof) of their proliferative potential. On the one hand, a school of thought suggests that chondroprogenitors have a limited proliferative potential that gets progressively exhausted with each cell division (5). In this scenario, catch-up growth is a cell-autonomous process: transient cell-cycle arrest leads to preservation of the proliferative potential in the arrested cells, so that these grow at a faster-than-normal-for-age rate once the insult is lifted. On the other hand, this model does not seem to be complete or even correct, as we have shown that mosaic (i.e. salt-an-pepper) cell-cycle arrest in the growth plate is compensated for by hyperproliferation of spared chondrocytes, which is a cell-nonautonomous response (7). Moreover, recent studies have shown that there is a change in the behavior of chondroprogenitors across development: from not self-renewing during fetal-neonatal stages, so that their pool is depleted as bones grow, to self-renewing and becoming long-lived progenitors or stem cells at juvenile stages, likely due to the influence of extrinsic signals, such as formation of the secondary ossification center (reviewed in (8)).
Oichi and colleagues shed light onto this process, by characterizing the fate, stem cell markers and growth potential of chondroprogenitors, as well as bone length and body weight, in mice exposed to three distinct regimes: ad libitum feeding (Normal), dietary restriction (DR), and dietary restriction followed by Refeeding (which causes catch-up growth). The preprint is very revealing, while opening new avenues for future research.
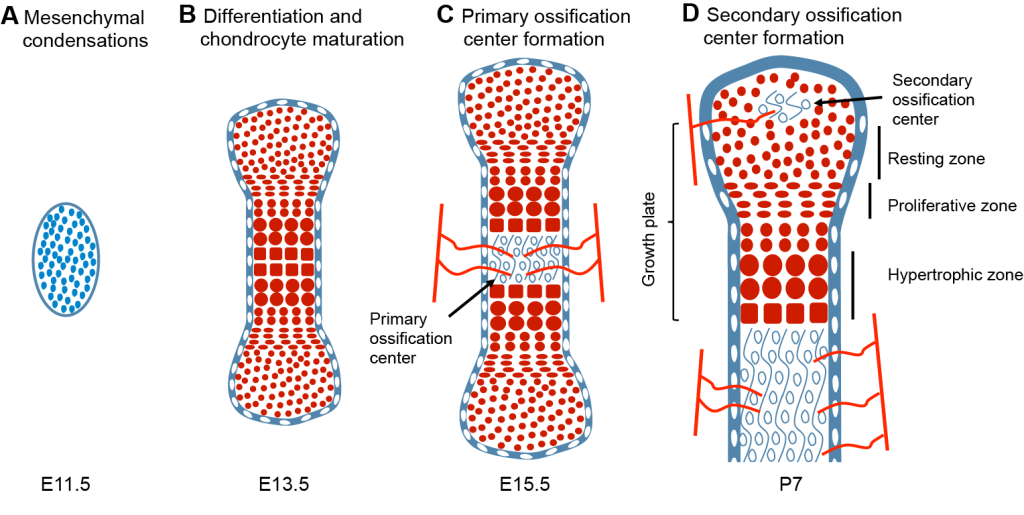
Figure 1: Process of endochondral ossification.
Key findings
Oichi et al. used lineage tracing of Axin2-CreER+ (Axin2+ hereafter) cells to trace the stem cells in the cartilage. They showed that the DR regime was able to stunt bone growth, as expected. Surprisingly, however, this was in part mediated by reduced recruitment (i.e. differentiation) of Axin2+ cells to the proliferative pool in the cartilage, and reciprocal increase of the resting pool (i.e. self-renewal, see Figure 2-middle).
- Following refeeding after nutrient restriction, the model demonstrated catch-up growth as evident by the increased growth rate as compared to age- and sex-matched Normal animals, leading to recovery of normal bone length a few weeks later. This correlated with increased differentiation of the Axin2+ cells towards the proliferative pool (Figure 2-right).
- Region-specific RNA-seq data and subsequent analysis revealed that IGF-1/PI3K signaling was differentially activated in resting vs. proliferative chondrocytes. Interestingly, IGF-1 signaling was downregulated during the DR regimen, correlating with increased self-renewal capacity and suppressed cell differentiation of the resting cells (Figure 2-middle). Of note, exogenous application of recombinant human (rh) IGF-1 was able to partially reverse this cell behavior.
- Conversely, refeeding after DR rescued IGF-1/PI3K signaling in resting chondrocytes, allowing the enhanced recruitment of the previously increased pool of resting cells to the proliferative pool, thus accelerating bone growth.
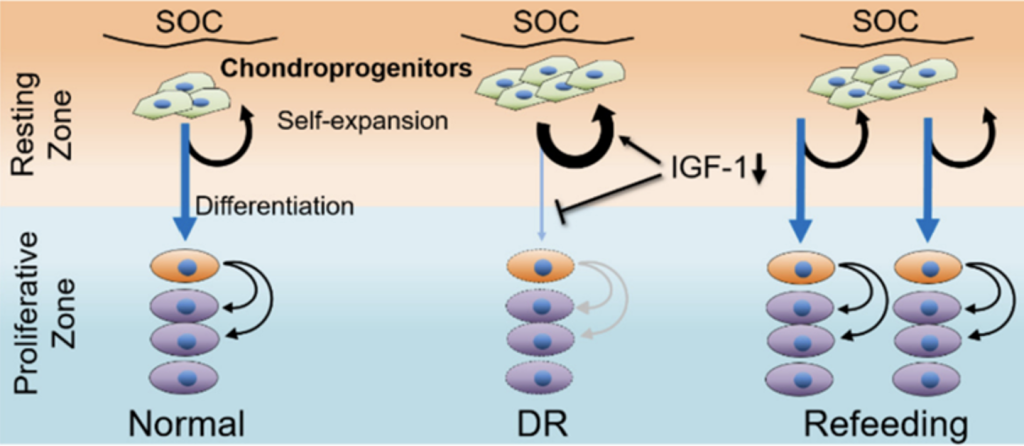
Figure 2: Summary of findings. Oichi et al. bioRxiv 2023.
What we like about this preprint
This study clearly shows that catch-up growth due to transient nutrient restriction is a two-step process: during DR, the Axin2+ resting cells “save energy” by reducing recruitment into the proliferative pool, preserving their proliferative potential. When the conditions are more favorable, this expanded pool of progenitors with high proliferative potential is the one driving enhanced growth as compared to Normal animals (which have fewer Axin2+ cells by that stage).
Moreover, one of the predictions of the cell-autonomous model of catch-up growth is that, when a bone is smaller than it should, given its chronological age, it will catch-up at a rate similar to that of a younger bone of similar size (which would be its ‘bone age’). Oichi and colleagues provide the exact data needed to test this prediction. In Figure S7, they show that tibial length at P41 in the Refeeding group is similar to tibial length in the P34 Normal group (this is the case for both males and females). A similar growth rate would then be expected for both groups. But this is not always the case. In males, P34 Normal growth rate is ~140 µm/day, but P41 Refeeding growth rate is ~100 µm/day, and the low variability suggests that this difference is significant. In females, however, P34 Normal growth rate is ~110 µm/day, very similar to P41 Refeeding growth rate, as predicted by the cell-autonomous model. The cause of this ‘sexual dimorphism’ remains to be determined, but it suggests that other factors, such as metabolism, hormones or body weight, can modulate the catch-up process.
Pending questions
– What about other chondroprogenitors, besides Axin2+ cells? Do they behave similarly?
– Newton et al. (9) showed that mTORC1 activity is required for the stem-cell behavior of self-renewing chondroprogenitors. However, Oichi and colleagues show that replenishment of the reserve pool is promoted by reduced IGF1 signaling, which is typically upstream of mTORC1. It seems that new experiments are required to reconcile these two observations.
– Fig 1e: Total Axin2+ cells were quantified apparently without normalization. What is the proportion of total Axin2+ cells referred to the total number of resting chondrocytes? In other words, is there a change in cell density?
– What happens if prolonged rhIGF-1 treatment is given? Do you expect to see overgrowth (increased differentiation, number and density of column) in the DR + rhIGF-1 chondrocytes?
Related research
- A. Prader, J. M. Tanner, G. von Harnack, Catch-up growth following illness or starvation. An example of developmental canalization in man. J Pediatr 62, 646-659 (1963).
- C. H. Waddington, The strategy of the genes; a discussion of some aspects of theoretical biology. (Allen & Unwin, London,, 1957), pp. ix, 262 p.
- I. I. Schmalhausen, Factors of evolution: the theory of stabilizing selection. Factors of evolution: the theory of stabilizing selection. (Blakiston, Oxford, England, 1949), pp. xiv, 327-xiv, 327.
- J. M. Tanner, Regulation of Growth in Size in Mammals. Nature 199, 845-850 (1963).
- J. Baron et al., Catch-up growth after glucocorticoid excess: a mechanism intrinsic to the growth plate. Endocrinology 135, 1367-1371 (1994).
- H. M. Kronenberg, Developmental regulation of the growth plate. Nature 423, 332-336 (2003).
- A. Rosello-Diez, L. Madisen, S. Bastide, H. Zeng, A. L. Joyner, Cell-nonautonomous local and systemic responses to cell arrest enable long-bone catch-up growth in developing mice. PLoS Biol 16, e2005086 (2018).
- J. C. Lui, Home for a rest: stem cell niche of the postnatal growth plate. J Endocrinol 246, R1-R11 (2020).
- P. T. Newton et al., A radical switch in clonality reveals a stem cell niche in the epiphyseal growth plate. Nature 567, 234-238 (2019).
doi: https://doi.org/10.1242/prelights.33860
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Cooperation between cortical and cytoplasmic forces shapes planar 4-cell stage embryos
Corentin Mollier, Shivani Dharmadhikari
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)