Primate naïve pluripotent stem cells stall in the G1 phase of the cell cycle and differentiate prematurely during embryo colonization
Posted on: 4 June 2020
Preprint posted on 28 March 2020
Article now published in Stem Cell Reports at http://dx.doi.org/10.1016/j.stemcr.2020.12.004
Mouse naïve pluripotent cells can colonise a distantly related species’ embryo, but why can’t primate PSCs do the same?
Selected by Pierre OsteilCategories: developmental biology
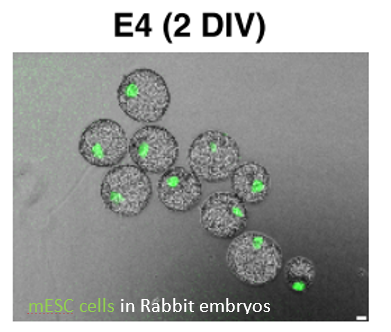
Background:
The first report on the capability of mouse embryonic stem cells (mESCs), derived from a blastocyst, to contribute to the development of an embryo when injected into the blastocyst (Day 3.5 post coitus) 1 opened a new era for functional genomics. Indeed, since then scientists can genetically engineer mESCs and create chimeras. If the modified ESCs contribute to the germ line, the next generation of animals will be 100% transgenic. From there, many projects originated with a similar aim of achieving a comparable outcome with other mammalian species, such as monkey, our closest relative. But for decades research has faced an important challenge: generating chimeras only works efficiently in mouse but not in other mammals. Interestingly, pluripotent stem cells can also be derived from mouse epiblast, termed Epiblast stem cells or EpiSC 4,5. However, these are incapable of contributing to the mouse embryo. This revealed the broad spectrum of pluripotency: the self-renewal status of pluripotent stem cells (PSCs) was coined “Naïve” when a PSC can contribute to the embryo to full term development or “Primed” for those that cannot 6.
From there, a new goal was set: make non-rodent mammalian PSCs “naïve” again!
So, a wide range of protocols emerged to reprogram primate PSCs into a state close to that of the mESC 7,8,17,9–16. Gene expression, colony morphologies as well as epigenetic features were used to grant them the status of naïve.
But are they really naïve? To tackle this question, Aksoy & colleagues decided to revisit different naïve conversion protocols, then see if the reprogramming was sufficient to acquire chimerism potential.
The results:
The rabbit embryo: an unnoticed model in the developmental and stem cell field
One number is striking: 2956 rabbit embryos have been used in total for chimera injection. Additionally, the rabbit embryo gastrulates as a flat disk, such as primate. The material abundance and the physical similarity of the rabbit embryos make this species a very adequate model for interspecies chimeras. On the other hand, only 36 cynomolgus monkey embryos were used, which, despite being a great achievement, demonstrates the challenges for large scale study on non-human primates.
First, Aksoy & colleagues injected mESCs into rabbit embryos from serum/LIF (n=238) and 2i/LIF (n=19). 98% of these embryos contained Serum/LIF GFP+ cells expressing NANOG and SOX2 but not SOX17, suggesting the cells are not incorporating the primitive endoderm layer but only the epiblast. This demonstrates that the rabbit embryo can be used for interspecies chimera studies.
Naïve non-human primate cells are not able to form chimeras
Then, they reprogrammed the primed Rhesus monkey PSCs using 7 different protocols that have been reported for successful conversion of the naïve cells. Together with the primed culture condition as a control, Rhesus PSCs were injected into 2385 embryos in total (so an average of about 300 embryos per condition). Four conditions showed successful incorporation after 3 days of culture: TL2i9, 4i/L/b16, T2iLGoY 15and LCDM (EPS)17. These conditions reprogrammed the rhPSC to a state comparable to that of the E6/E7 cynomolgus epiblast according to the transcriptome analysis (see the PCA below). Despite incorporation, they were not able to survive and divide after 3 days in the embryo. TL2i cells show the highest survival rate of 57% and seem to display an increased cell number at day 2 despite the premature expression of Gata6, a primitive endoderm marker. Even ROCK inhibitor did not rescue the survival. Similar results were obtained with human iPSCs grown in TL2i.
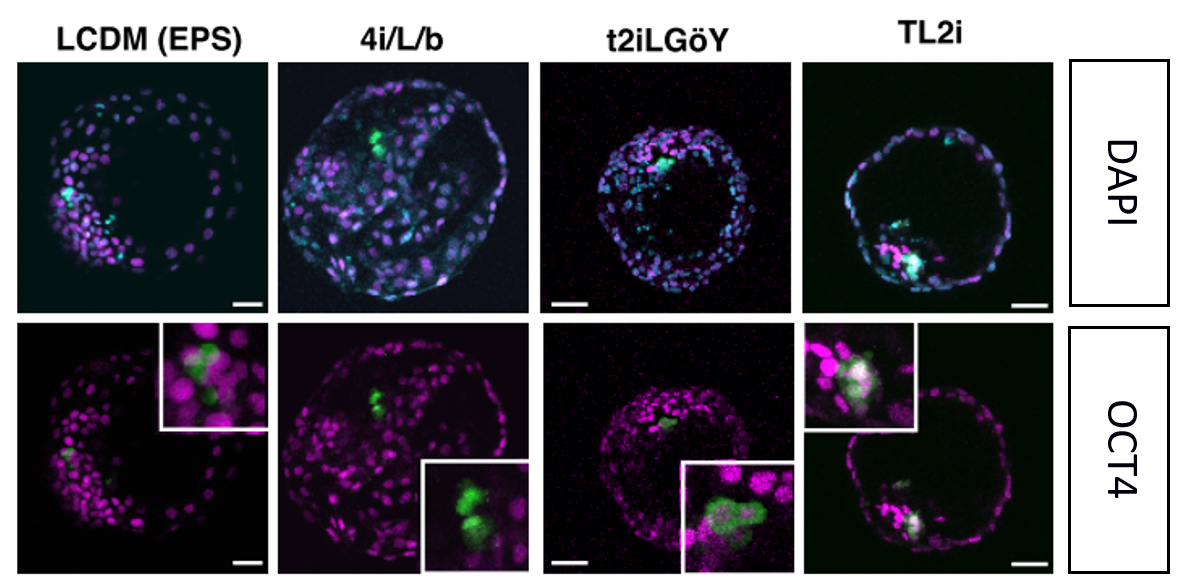
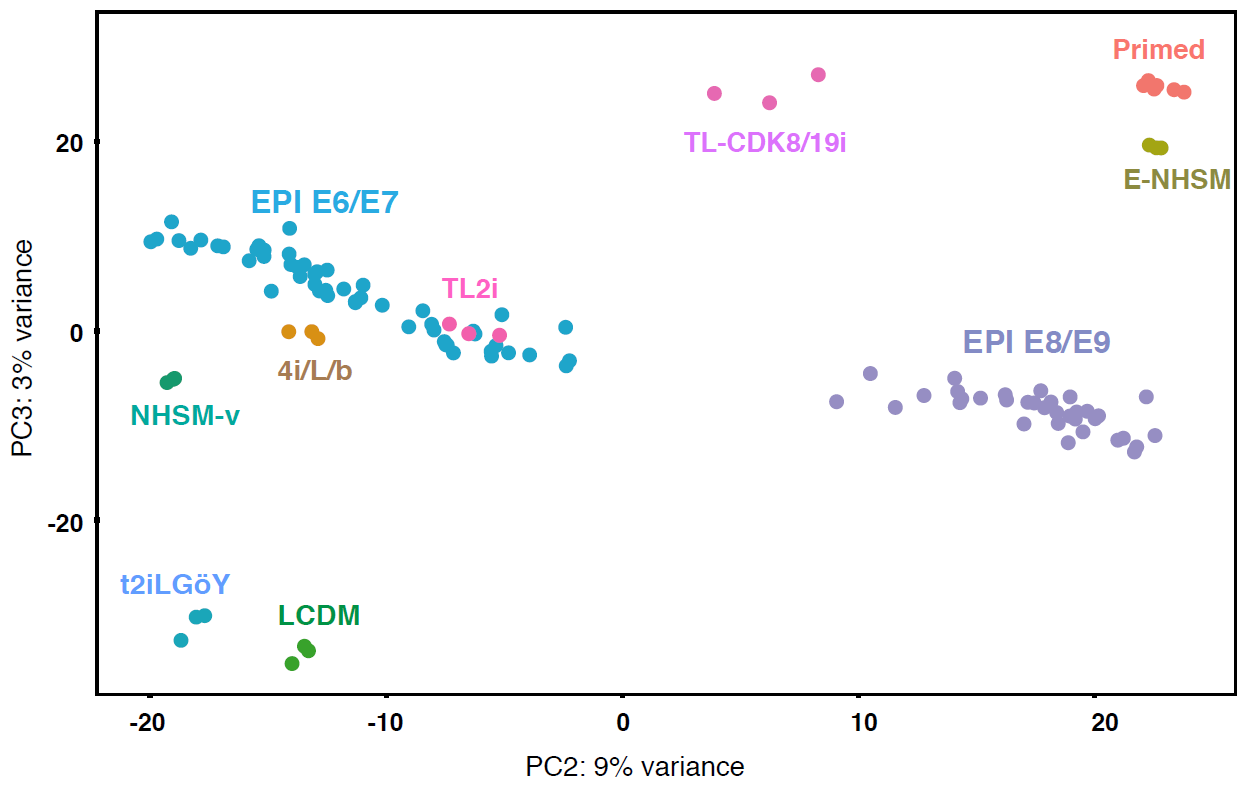
The low contribution of primate PSCs might be due to the failure to proliferate when in single cell suspension.
After obtaining these results, Aksoy & colleagues decided to answer the question of whether this incapability of incorporation into the blastocyst was due to evolutionary distance. So, they injected rhesus TL2i and human t2iLGoY into cynomolgus embryos (7 and 15 respectively) while 5 were injected with mESCs. Similar results as with the rabbit embryos were found suggesting the evolutionary distance between rabbit and rhesus monkey is not the key factor here, since mESCs can contribute.
Then, they investigated DNA replication in a condition similar to that of injection into the embryo (here DNA replication after single cell dissociation). mESCs do not show any changes in DNA replication. But for the monkey ESCs the story is quite different. First, DNA replication is significantly slower in rhESCs, but by 1 hour after dissociation proliferative cells are almost inexistent (see FACS plots) suggesting that the non-human PSCs do not replicate their DNA while in single-cell suspension. When reprogrammed into naïve condition (4i/L/b, TL2i and t2iLGoY) cells had an increased DNA replication rate but this was not maintained after dissociation. This was confirmed in a chimera set up, where only 4 cells out of 29 still replicates 24hours after injection.
Non-human primate naïve cells stop proliferating in G1.
FUCCI mESCs and rhESCs were generated. The team observed that the distribution is quite different with an increased G1 phase in rhesus cells of 43% compared to 18% in mESCs. After conversion of the rhPSCs into naïve condition cells and their injection into rabbit embryos almost all the cells (78% in 4i/L/b and 100% in TL2i) are stuck in G1, suggesting growth arrest.
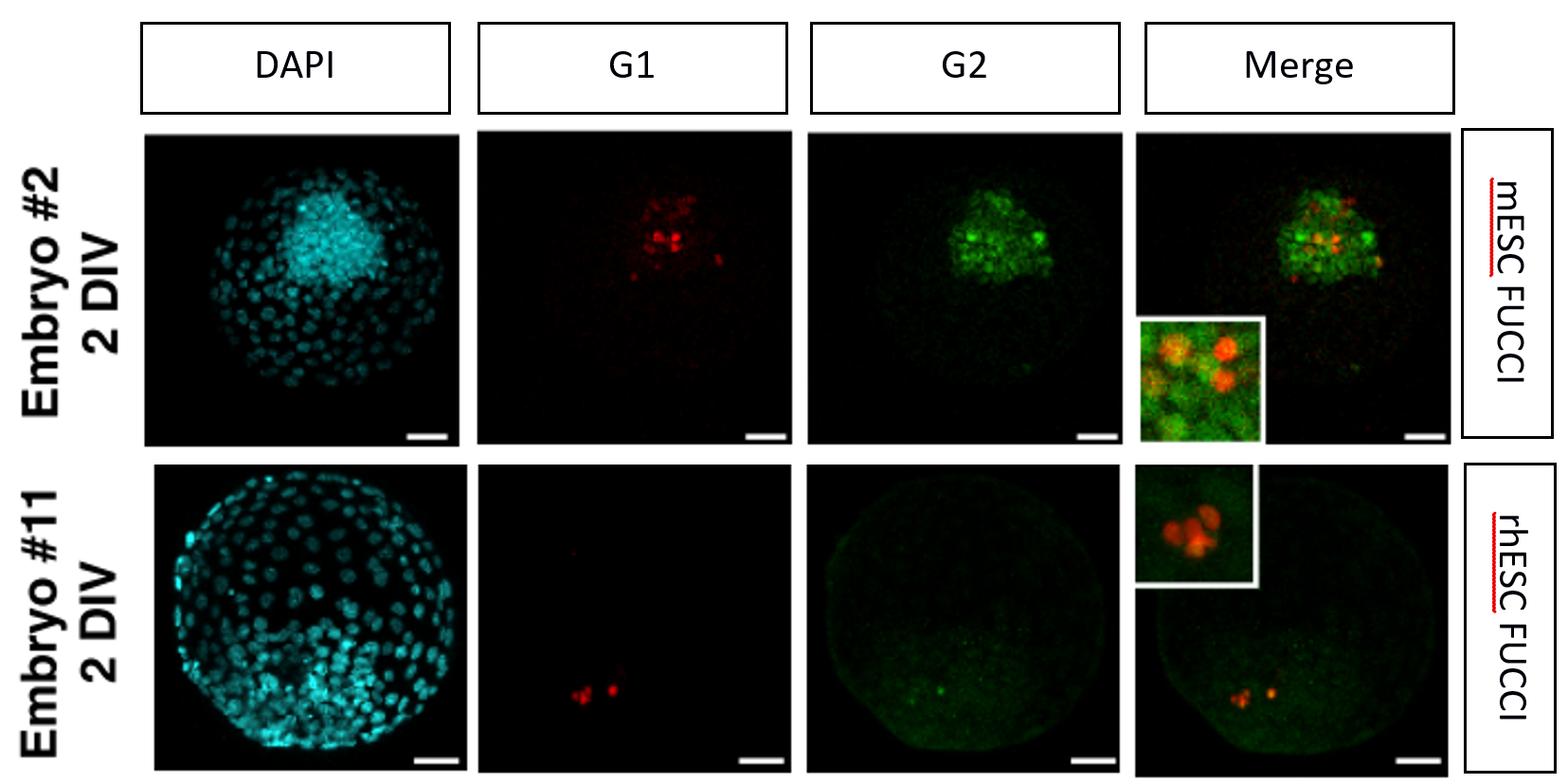
Conclusion:
The reprogramming of the rhPSCs into the so-called naïve state does not restore chimerism potential which is likely due to the culture conditions not supporting cell cycle progression after dissociation prior to blastocyst injection.
My take on this preprint:
I decided to cover this article as I did my PhD in this lab, so I am quite passionate by the questions tackled by my former team. The power of the research conducted herein lies in the ability to study multiple mammalian species together: mouse, rabbit, monkey and human. This is a first-of-its-kind study trying to solve the question of whether multiple species’ naïve pluripotent cells are able to colonise a blastocyst. But this is not the case suggesting we might need to redefine what is called “naïve” pluripotency. The large number of embryos used in this study is making a strong case for the need to reinvestigate pluripotency of non-human primate embryos.
4 questions to the authors:
1- At the end of the first result paragraph: “After gastrulation, mESCs were able to contribute only to the neuroectoderm, but not to other ectodermal structures, or to the mesoderm and endoderm of rabbit gastrula”. I found this particularly interesting. To me it means that mESCs, despite the fact that they survive and divide in rabbit embryos, are excluded from the primitive streak and for me “fail” to gastrulate. They thrive in the epiblast that becomes neurectoderm. What is your opinion on this result?
2- It doesn’t seem that you describe how you removed the autofluorescence from the imaging. “To overcome this limitation, we systematically used an anti- GFP antibody.” I don’t understand what you have done here? On supp figure 1 you show some imaging with the antibody without staining, but how did you manage to remove the autofluorescence?
3- Wouldn’t you agree that the TL2i is the best strategy so far for interspecies chimera potential? On this note, on p12 you said “At 3 DIV, 0% (n = 6),0% (n = 4), and 50% (n = 2)” figure 5D shows 80%, 80%, and 20% of HuN cells positive for SOX2, NANOG and GATA6”. If it is true, TL2i leads to more contribution to the epiblast lineage compare t2iLGoY (33% of cells into the primitive endoderm) which is supported by the PCA showing TL2i overlap with epiblast cells.
4- One last question: Do you think your rabbit, monkey and human PSC are naïve?
References:
- Bradley, A., Evans, M., Kaufman, M. H. & Robertson, E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255–256 (1984).
- Tachibana, M. et al. Generation of chimeric rhesus monkeys. Cell 148, 285–295 (2012).
- Li, P. et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell 135, 1299–310 (2008).
- Brons, I. G. et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 (2007).
- Tesar, P. J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007).
- Nichols, J. & Smith, A. Naive and primed pluripotent states. Cell Stem Cell 4, 487–92 (2009).
- Gafni, O. et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286 (2013).
- Gao, X. et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 21, 687–699 (2019).
- Chen, H. et al. Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat. Commun. 6, 7095 (2015).
- Takashima, Y. et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell 162, 452–453 (2015).
- Chan, Y. S. et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13, 663–675 (2013).
- Theunissen, T. W. et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 471–487 (2014).
- Ohtsuka, S., Nishikawa-Torikai, S. & Niwa, H. E-cadherin promotes incorporation of mouse epiblast stem cells into normal development. PLoS One 7, e45220 (2012).
- Guo, G. et al. Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Reports 6, 437–446 (2016).
- Guo, G. et al. Epigenetic resetting of human pluripotency. Development 145, (2018).
- Fang, R. et al. Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell 15, 488–497 (2014).
- Yang, J. et al. Establishment of mouse expanded potential stem cells. Nature 550, 393–397 (2017).
doi: https://doi.org/10.1242/prelights.21571
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Cooperation between cortical and cytoplasmic forces shapes planar 4-cell stage embryos
Corentin Mollier, Shivani Dharmadhikari
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)